1WMX
 
 | | Crystal Structure of Family 30 Carbohydrate Binding Module | | Descriptor: | COG3291: FOG: PKD repeat, SULFATE ION | | Authors: | Horiguchi, Y, Kono, M, Suzuki, A, Yamane, T, Arai, M, Sakka, K, Omiya, K. | | Deposit date: | 2004-07-21 | | Release date: | 2004-08-03 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of Family 30 Carbohydrate Binding Module
To be Published
|
|
1WZX
 
 | | Crystal Structure of Family 30 Carbohydrate Binding Module. | | Descriptor: | COG3291: FOG: PKD repeat | | Authors: | Horiguchi, Y, Kono, M, Suzuki, A, Yamane, T, Arai, M, Sakka, K, Omiya, K. | | Deposit date: | 2005-03-10 | | Release date: | 2005-03-22 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3.52 Å) | | Cite: | Crystal Structure of Family 30 Carbohydrate Binding Module
To be Published
|
|
1VEJ
 
 | | Solution Structure of RSGI RUH-016, a UBA Domain from mouse cDNA | | Descriptor: | RIKEN cDNA 4931431F19 | | Authors: | Higuchi, Y, Abe, T, Hirota, H, Hayashi, F, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2004-03-31 | | Release date: | 2005-05-31 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of RSGI RUH-016, a UBA Domain from mouse cDNA
To be Published
|
|
1VEK
 
 | | Solution Structure of RSGI RUH-011, a UBA Domain from Arabidopsis cDNA | | Descriptor: | ubiquitin-specific protease 14, putative | | Authors: | Higuchi, Y, Abe, T, Hirota, H, Saito, K, Koshiba, S, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2004-03-31 | | Release date: | 2004-09-30 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of RSGI RUH-011, a UBA Domain from Arabidopsis cDNA
To be Published
|
|
7E9U
 
 | | Trehalase of Arabidopsis thaliana | | Descriptor: | GLYCEROL, PHOSPHATE ION, SODIUM ION, ... | | Authors: | Taguchi, Y, Saburi, W, Yu, J, Imai, R, Yao, M, Mori, H. | | Deposit date: | 2021-03-05 | | Release date: | 2022-03-09 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | pH-dependent alteration of substrate specificity of plant trehalase and its molecular mechanism
To Be Published
|
|
7E9X
 
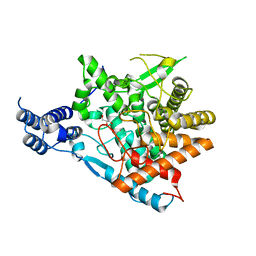 | | Trehalase of Arabidopsis thaliana acid mutant -D380A | | Descriptor: | GLYCEROL, Trehalase | | Authors: | Taguchi, Y, Saburi, W, Yu, J, Imai, R, Yao, M, Mori, H. | | Deposit date: | 2021-03-05 | | Release date: | 2022-03-09 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | pH-dependent alteration of substrate specificity of plant trehalase and its molecular mechanism
To Be Published
|
|
7EAW
 
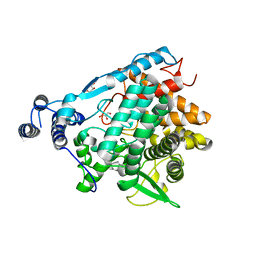 | | Trehalase of Arabidopsis thaliana acid mutant -D380A trehalose complex | | Descriptor: | GLYCEROL, Trehalase, alpha-D-glucopyranose-(1-1)-alpha-D-glucopyranose | | Authors: | Taguchi, Y, Saburi, W, Yu, J, Imai, R, Yao, M, Mori, H. | | Deposit date: | 2021-03-08 | | Release date: | 2022-03-16 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | pH-dependent alteration of substrate specificity of plant trehalase and its molecular mechanism
To Be Published
|
|
6IUJ
 
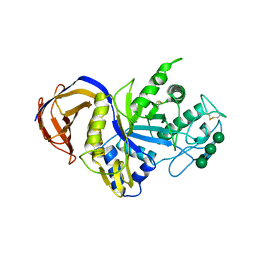 | | Crystal structure of GH30 xylanase B from Talaromyces cellulolyticus | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, GH30 Xylanase B, ... | | Authors: | Nakamichi, Y, Watanabe, M, Inoue, H. | | Deposit date: | 2018-11-28 | | Release date: | 2019-01-30 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structural and functional characterization of a bifunctional GH30-7 xylanase B from the filamentous fungusTalaromyces cellulolyticus.
J. Biol. Chem., 294, 2019
|
|
2ZWO
 
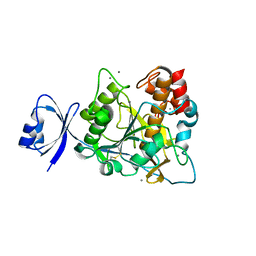 | | Crystal structure of Ca2 site mutant of Pro-S324A | | Descriptor: | CALCIUM ION, Tk-subtilisin | | Authors: | Takeuchi, Y, Tanaka, S, Matsumura, H, Koga, Y, Takano, K, Kanaya, S. | | Deposit date: | 2008-12-17 | | Release date: | 2009-06-23 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Requirement of a unique Ca(2+)-binding loop for folding of Tk-subtilisin from a hyperthermophilic archaeon.
Biochemistry, 48, 2009
|
|
2ZWP
 
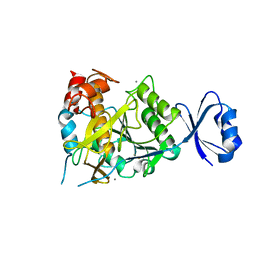 | | Crystal structure of Ca3 site mutant of Pro-S324A | | Descriptor: | CALCIUM ION, Tk-subtilisin | | Authors: | Takeuchi, Y, Tanaka, S, Matsumura, H, Koga, Y, Takano, K, Kanaya, S. | | Deposit date: | 2008-12-17 | | Release date: | 2009-06-23 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Requirement of a unique Ca(2+)-binding loop for folding of Tk-subtilisin from a hyperthermophilic archaeon.
Biochemistry, 48, 2009
|
|
3WYH
 
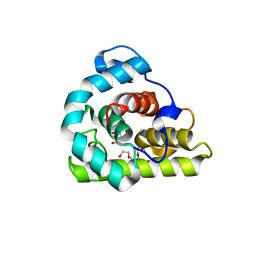 | |
2ZJD
 
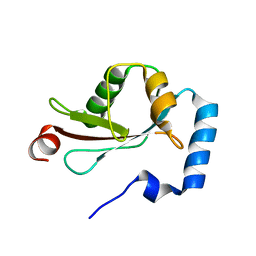 | | Crystal Structure of LC3-p62 complex | | Descriptor: | Microtubule-associated proteins 1A/1B light chain 3B precursor, undecameric peptide from Sequestosome-1 | | Authors: | Ichimura, Y, Kumanomidou, T, Sou, Y, Mizushima, T, Ezaki, J, Ueno, T, Kominami, E, Yamane, T, Tanaka, K, Komatsu, M. | | Deposit date: | 2008-03-05 | | Release date: | 2008-06-03 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | Structural Basis for Sorting Mechanism of p62 in Selective Autophagy
J.Biol.Chem., 283, 2008
|
|
2ZJW
 
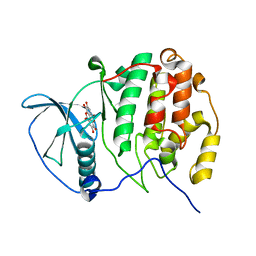 | | Crystal structure of human CK2 alpha complexed with Ellagic acid | | Descriptor: | 2,3,7,8-tetrahydroxychromeno[5,4,3-cde]chromene-5,10-dione, Casein kinase II subunit alpha | | Authors: | Sekiguchi, Y, Kinoshita, T, Nakaniwa, T, Tada, T. | | Deposit date: | 2008-03-11 | | Release date: | 2009-03-17 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural insight into human CK2alpha in complex with the potent inhibitor ellagic acid
Bioorg.Med.Chem.Lett., 19, 2009
|
|
3AQO
 
 | |
5B1N
 
 | | DHp domain structure of EnvZ from Escherichia coli | | Descriptor: | Osmolarity sensor protein EnvZ | | Authors: | Okajima, T, Eguchi, Y, Tochio, N, Inukai, Y, Shimizu, R, Ueda, S, Shinya, S, Kigawa, T, Fukamizo, T, Igarashi, M, Utsumi, R. | | Deposit date: | 2015-12-09 | | Release date: | 2016-12-14 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.33 Å) | | Cite: | Angucycline antibiotic waldiomycin recognizes common structural motif conserved in bacterial histidine kinases
J. Antibiot., 70, 2017
|
|
5AX7
 
 | | yeast pyruvyltransferase Pvg1p | | Descriptor: | Pyruvyl transferase 1, ZINC ION | | Authors: | Kanekiyo, M, Yoritsune, K, Yoshinaga, S, Higuchi, Y, Takegawa, K, Kakuta, Y. | | Deposit date: | 2015-07-16 | | Release date: | 2016-06-08 | | Last modified: | 2020-02-26 | | Method: | X-RAY DIFFRACTION (2.46 Å) | | Cite: | A rationally engineered yeast pyruvyltransferase Pvg1p introduces sialylation-like properties in neo-human-type complex oligosaccharide
Sci Rep, 6, 2016
|
|
3AMY
 
 | | Crystal structure of human CK2 alpha complexed with apigenin | | Descriptor: | 1,2-ETHANEDIOL, 5,7-dihydroxy-2-(4-hydroxyphenyl)-4H-chromen-4-one, Casein kinase II subunit alpha | | Authors: | Sekiguchi, Y, Nakaniwa, T, Kinoshita, T, Tada, T. | | Deposit date: | 2010-08-25 | | Release date: | 2011-10-05 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of human CK2 alpha complexed with apigenin
To be Published
|
|
5SIC
 
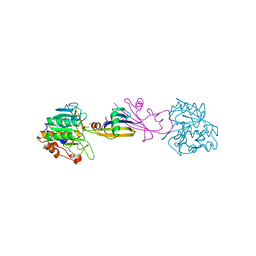 | |
6KBR
 
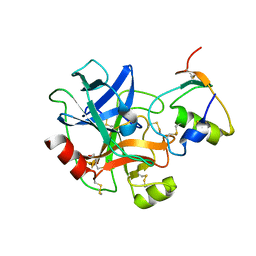 | |
6KRU
 
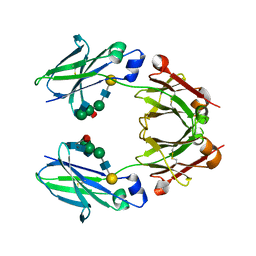 | | Crystal structure of mouse IgG2b Fc | | Descriptor: | Ig gamma-2B chain C region, beta-D-galactopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)-[2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)]alpha-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, beta-D-galactopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)-[2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)]alpha-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Taniguchi, Y, Satoh, T, Yagi, H, Kato, K. | | Deposit date: | 2019-08-22 | | Release date: | 2020-01-22 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | On-Membrane Dynamic Interplay between Anti-GM1 IgG Antibodies and Complement Component C1q.
Int J Mol Sci, 21, 2019
|
|
6KRV
 
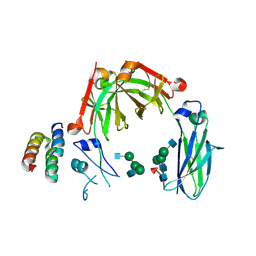 | | Crystal structure of mouse IgG2b Fc complexed with B domain of Protein A | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)]alpha-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)]alpha-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, Ig gamma-2B chain C region, ... | | Authors: | Taniguchi, Y, Satoh, T, Yagi, H, Kato, K. | | Deposit date: | 2019-08-22 | | Release date: | 2020-01-22 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | On-Membrane Dynamic Interplay between Anti-GM1 IgG Antibodies and Complement Component C1q.
Int J Mol Sci, 21, 2019
|
|
6LVL
 
 | | Crystal structure of FGFR2 in complex with 1,3,5-triazine derivative | | Descriptor: | Fibroblast growth factor receptor 2, N-ethyl-2-[[4-[[3-methoxy-4-[4-(4-methylpiperazin-1-yl)piperidin-1-yl]phenyl]amino]-1,3,5-triazin-2-yl]amino]benzenesulfonamide, SULFATE ION | | Authors: | Echizen, Y, Tateishi, Y, Amano, Y. | | Deposit date: | 2020-02-04 | | Release date: | 2020-04-08 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.98 Å) | | Cite: | Structure-based drug design of 1,3,5-triazine and pyrimidine derivatives as novel FGFR3 inhibitors with high selectivity over VEGFR2.
Bioorg.Med.Chem., 28, 2020
|
|
6LVM
 
 | | Crystal structure of FGFR3 in complex with pyrimidine derivative | | Descriptor: | 2-[[5-[2-(3,5-dimethoxyphenyl)ethyl]-2-[[3-methoxy-4-[4-(4-methylpiperazin-1-yl)piperidin-1-yl]phenyl]amino]pyrimidin-4-yl]amino]-N-ethyl-benzenesulfonamide, Fibroblast growth factor receptor 3 | | Authors: | Echizen, Y, Tateishi, Y, Amano, Y. | | Deposit date: | 2020-02-04 | | Release date: | 2020-04-08 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.53 Å) | | Cite: | Structure-based drug design of 1,3,5-triazine and pyrimidine derivatives as novel FGFR3 inhibitors with high selectivity over VEGFR2.
Bioorg.Med.Chem., 28, 2020
|
|
6LVK
 
 | | Crystal structure of FGFR2 in complex with 1,3,5-triazine derivative | | Descriptor: | Fibroblast growth factor receptor 2, N-ethyl-2-[[4-[[4-(4-methylpiperazin-1-yl)-3-(2-morpholin-4-ylethoxy)phenyl]amino]-1,3,5-triazin-2-yl]amino]benzenesulfonamide, SULFATE ION | | Authors: | Echizen, Y, Amano, Y, Tateishi, Y. | | Deposit date: | 2020-02-04 | | Release date: | 2020-04-08 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | Structure-based drug design of 1,3,5-triazine and pyrimidine derivatives as novel FGFR3 inhibitors with high selectivity over VEGFR2.
Bioorg.Med.Chem., 28, 2020
|
|
5B1O
 
 | | DHp domain structure of EnvZ P248A mutant | | Descriptor: | Osmolarity sensor protein EnvZ | | Authors: | Okajima, T, Eguchi, Y, Tochio, N, Inukai, Y, Shimizu, R, Ueda, S, Shinya, S, Kigawa, T, Fukamizo, T, Igarashi, M, Utsumi, R. | | Deposit date: | 2015-12-09 | | Release date: | 2016-12-14 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Angucycline antibiotic waldiomycin recognizes common structural motif conserved in bacterial histidine kinases
J. Antibiot., 70, 2017
|
|
