1Q6E
 
 | | Crystal Structure of Soybean Beta-Amylase Mutant (E178Y) with Increased pH Optimum at pH 5.4 | | Descriptor: | SULFATE ION, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose, alpha-D-glucopyranose-(1-4)-beta-D-glucopyranose, ... | | Authors: | Hirata, A, Adachi, M, Sekine, A, Kang, Y.N, Utsumi, S, Mikami, B. | | Deposit date: | 2003-08-13 | | Release date: | 2004-02-24 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural and Enzymatic Analysis of Soybean {beta}-Amylase Mutants with Increased pH Optimum
J.Biol.Chem., 279, 2004
|
|
1Q6G
 
 | | Crystal Structure of Soybean Beta-Amylase Mutant (N340T) with Increased pH Optimum | | Descriptor: | SULFATE ION, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose, alpha-D-glucopyranose-(1-4)-beta-D-glucopyranose, ... | | Authors: | Hirata, A, Adachi, M, Sekine, A, Kang, Y.N, Utsumi, S, Mikami, B. | | Deposit date: | 2003-08-13 | | Release date: | 2004-02-24 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural and Enzymatic Analysis of Soybean {beta}-Amylase Mutants with Increased pH Optimum
J.Biol.Chem., 279, 2004
|
|
1Q6F
 
 | | Crystal Structure of Soybean Beta-Amylase Mutant (E178Y) with Increased pH Optimum at pH 7.1 | | Descriptor: | SULFATE ION, alpha-D-glucopyranose-(1-4)-beta-D-glucopyranose, beta-D-glucopyranose-(1-4)-beta-D-glucopyranose, ... | | Authors: | Hirata, A, Adachi, M, Sekine, A, Kang, Y.N, Utsumi, S, Mikami, B. | | Deposit date: | 2003-08-13 | | Release date: | 2004-02-24 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural and Enzymatic Analysis of Soybean {beta}-Amylase Mutants with Increased pH Optimum
J.Biol.Chem., 279, 2004
|
|
5X23
 
 | | Crystal structure of CYP2C9 genetic variant A477T (*30) in complex with multiple losartan molecules | | Descriptor: | Cytochrome P450 2C9, PHOSPHATE ION, POTASSIUM ION, ... | | Authors: | Maekawa, K, Adachi, M, Shah, M.B. | | Deposit date: | 2017-01-30 | | Release date: | 2017-10-25 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Basis of Single-Nucleotide Polymorphisms in Cytochrome P450 2C9
Biochemistry, 56, 2017
|
|
5X24
 
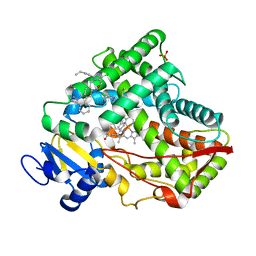 | | Crystal structure of CYP2C9 genetic variant I359L (*3) in complex with multiple losartan molecules | | Descriptor: | Cytochrome P450 2C9, PHOSPHATE ION, POTASSIUM ION, ... | | Authors: | Maekawa, K, Adachi, M, Shah, M.B. | | Deposit date: | 2017-01-30 | | Release date: | 2017-10-25 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | Structural Basis of Single-Nucleotide Polymorphisms in Cytochrome P450 2C9
Biochemistry, 56, 2017
|
|
5XXI
 
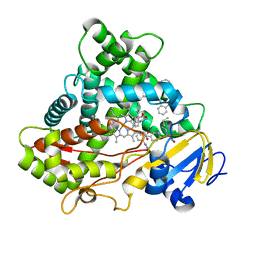 | | Crystal structure of CYP2C9 in complex with multiple losartan molecules | | Descriptor: | Cytochrome P450 2C9, POTASSIUM ION, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Maekawa, K, Adachi, M, Shah, M.B. | | Deposit date: | 2017-07-04 | | Release date: | 2017-10-25 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural Basis of Single-Nucleotide Polymorphisms in Cytochrome P450 2C9
Biochemistry, 56, 2017
|
|
1V3H
 
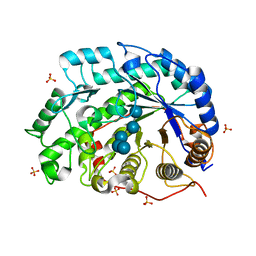 | | The roles of Glu186 and Glu380 in the catalytic reaction of soybean beta-amylase | | Descriptor: | Beta-amylase, SULFATE ION, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Kang, Y.N, Adachi, M, Utsumi, S, Mikami, B. | | Deposit date: | 2003-11-02 | | Release date: | 2004-06-22 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The Roles of Glu186 and Glu380 in the Catalytic Reaction of Soybean beta-Amylase.
J.Mol.Biol., 339, 2004
|
|
1UKP
 
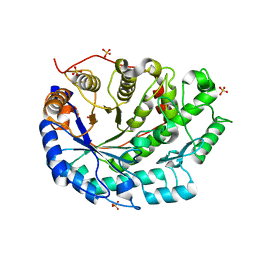 | | Crystal structure of soybean beta-amylase mutant substituted at surface region | | Descriptor: | Beta-amylase, SULFATE ION | | Authors: | Kang, Y.N, Adachi, M, Mikami, B, Utsumi, S. | | Deposit date: | 2003-08-31 | | Release date: | 2004-02-10 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Change in the crystal packing of soybean beta-amylase mutants substituted at a few surface amino acid residues
Protein Eng., 16, 2003
|
|
1UKO
 
 | | Crystal structure of soybean beta-amylase mutant substituted at surface region | | Descriptor: | Beta-amylase, SULFATE ION | | Authors: | Kang, Y.N, Adachi, M, Mikami, B, Utsumi, S. | | Deposit date: | 2003-08-30 | | Release date: | 2004-02-10 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Change in the crystal packing of soybean beta-amylase mutants substituted at a few surface amino acid residues
Protein Eng., 16, 2003
|
|
1V3I
 
 | | The roles of Glu186 and Glu380 in the catalytic reaction of soybean beta-amylase | | Descriptor: | Beta-amylase, SULFATE ION, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose, ... | | Authors: | Kang, Y.N, Adachi, M, Utsumi, S, Mikami, B. | | Deposit date: | 2003-11-02 | | Release date: | 2004-06-22 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The Roles of Glu186 and Glu380 in the Catalytic Reaction of Soybean beta-Amylase.
J.Mol.Biol., 339, 2004
|
|
1VEO
 
 | | Crystal Structure Analysis of Y164F/maltose of Bacillus cereus Beta-Amylase at pH 4.6 | | Descriptor: | Beta-amylase, CALCIUM ION, alpha-D-glucopyranose, ... | | Authors: | Hirata, A, Adachi, M, Utsumi, S, Mikami, B. | | Deposit date: | 2004-04-03 | | Release date: | 2005-05-24 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | Engineering of the pH optimum of Bacillus cereus beta-amylase: conversion of the pH optimum from a bacterial type to a higher-plant type
Biochemistry, 43, 2004
|
|
1VEP
 
 | | Crystal Structure Analysis of Triple (T47M/Y164E/T328N)/maltose of Bacillus cereus Beta-Amylase at pH 6.5 | | Descriptor: | Beta-amylase, CALCIUM ION, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose, ... | | Authors: | Hirata, A, Adachi, M, Utsumi, S, Mikami, B. | | Deposit date: | 2004-04-03 | | Release date: | 2005-05-24 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Engineering of the pH optimum of Bacillus cereus beta-amylase: conversion of the pH optimum from a bacterial type to a higher-plant type
Biochemistry, 43, 2004
|
|
1VEM
 
 | | Crystal Structure Analysis of Bacillus Cereus Beta-Amylase at the optimum pH (6.5) | | Descriptor: | Beta-amylase, CALCIUM ION, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Hirata, A, Adachi, M, Utsumi, S, Mikami, B. | | Deposit date: | 2004-04-03 | | Release date: | 2005-05-24 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Engineering of the pH optimum of Bacillus cereus beta-amylase: conversion of the pH optimum from a bacterial type to a higher-plant type
Biochemistry, 43, 2004
|
|
1VEN
 
 | | Crystal Structure Analysis of Y164E/maltose of Bacilus cereus Beta-amylase at pH 4.6 | | Descriptor: | Beta-amylase, CALCIUM ION, alpha-D-glucopyranose | | Authors: | Hirata, A, Adachi, M, Utsumi, S, Mikami, B. | | Deposit date: | 2004-04-03 | | Release date: | 2005-05-24 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Engineering of the pH optimum of Bacillus cereus beta-amylase: conversion of the pH optimum from a bacterial type to a higher-plant type
Biochemistry, 43, 2004
|
|
1WDS
 
 | | The role of an inner loop in the catalytic mechanism of soybean beta-amylase | | Descriptor: | Beta-amylase, SULFATE ION, alpha-D-glucopyranose, ... | | Authors: | Kang, Y.N, Adachi, M, Utsumi, S, Mikami, B. | | Deposit date: | 2004-05-17 | | Release date: | 2005-04-05 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Structural analysis of threonine 342 mutants of soybean beta-amylase: role of a conformational change of the inner loop in the catalytic mechanism.
Biochemistry, 44, 2005
|
|
1WDP
 
 | | The role of an inner loop in the catalytic mechanism of soybean beta-amylase | | Descriptor: | Beta-amylase, SULFATE ION | | Authors: | Kang, Y.N, Adachi, M, Utsumi, S, Mikami, B. | | Deposit date: | 2004-05-17 | | Release date: | 2005-04-05 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.27 Å) | | Cite: | Structural analysis of threonine 342 mutants of soybean beta-amylase: role of a conformational change of the inner loop in the catalytic mechanism.
Biochemistry, 44, 2005
|
|
1WDR
 
 | | The role of an inner loop in the catalytic mechanism of soybean beta-amylase | | Descriptor: | Beta-amylase, SULFATE ION, alpha-D-glucopyranose, ... | | Authors: | Kang, Y.N, Adachi, M, Utsumi, S, Mikami, B. | | Deposit date: | 2004-05-17 | | Release date: | 2005-04-05 | | Last modified: | 2021-11-10 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Structural analysis of threonine 342 mutants of soybean beta-amylase: role of a conformational change of the inner loop in the catalytic mechanism.
Biochemistry, 44, 2005
|
|
1WDQ
 
 | | The role of an inner loop in the catalytic mechanism of soybean beta-amylase | | Descriptor: | Beta-amylase, SULFATE ION, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Kang, Y.N, Adachi, M, Utsumi, S, Mikami, B. | | Deposit date: | 2004-05-17 | | Release date: | 2005-04-05 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.28 Å) | | Cite: | Structural analysis of threonine 342 mutants of soybean beta-amylase: role of a conformational change of the inner loop in the catalytic mechanism.
Biochemistry, 44, 2005
|
|
8J6G
 
 | |
3HGP
 
 | | Structure of porcine pancreatic elastase complexed with a potent peptidyl inhibitor FR130180 determined by high resolution crystallography | | Descriptor: | 4-[[(2S)-3-methyl-1-oxo-1-[(2S)-2-[[(3S)-1,1,1-trifluoro-4-methyl-2-oxo-pentan-3-yl]carbamoyl]pyrrolidin-1-yl]butan-2-yl]carbamoyl]benzoic acid, CALCIUM ION, Elastase-1, ... | | Authors: | Tamada, T, Kinoshita, T, Kuroki, R, Tada, T. | | Deposit date: | 2009-05-14 | | Release date: | 2009-07-28 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (0.94 Å) | | Cite: | Combined High-Resolution Neutron and X-ray Analysis of Inhibited Elastase Confirms the Active-Site Oxyanion Hole but Rules against a Low-Barrier Hydrogen Bond
J.Am.Chem.Soc., 131, 2009
|
|
3HGN
 
 | | Structure of porcine pancreatic elastase complexed with a potent peptidyl inhibitor FR130180 determined by neutron crystallography | | Descriptor: | 4-[[(2S)-3-methyl-1-oxo-1-[(2S)-2-[[(3S)-1,1,1-trifluoro-4-methyl-2-oxo-pentan-3-yl]carbamoyl]pyrrolidin-1-yl]butan-2-yl]carbamoyl]benzoic acid, CALCIUM ION, Elastase-1, ... | | Authors: | Tamada, T, Kinoshita, T, Kuroki, R, Tada, T. | | Deposit date: | 2009-05-14 | | Release date: | 2009-07-28 | | Last modified: | 2023-11-01 | | Method: | NEUTRON DIFFRACTION (1.65 Å), X-RAY DIFFRACTION | | Cite: | Combined High-Resolution Neutron and X-ray Analysis of Inhibited Elastase Confirms the Active-Site Oxyanion Hole but Rules against a Low-Barrier Hydrogen Bond
J.Am.Chem.Soc., 131, 2009
|
|
5EVY
 
 | | Salicylate hydroxylase substrate complex | | Descriptor: | 2-HYDROXYBENZOIC ACID, FLAVIN-ADENINE DINUCLEOTIDE, Salicylate hydroxylase | | Authors: | Morimoto, Y, Uemura, T. | | Deposit date: | 2015-11-20 | | Release date: | 2015-12-16 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.47 Å) | | Cite: | The catalytic mechanism of decarboxylative hydroxylation of salicylate hydroxylase revealed by crystal structure analysis at 2.5 angstrom resolution
Biochem.Biophys.Res.Commun., 469, 2016
|
|
3WBH
 
 | | Structural characteristics of alkaline phosphatase from a moderately halophilic bacteria Halomonas sp.593 | | Descriptor: | Alkaline phosphatase, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Arai, S, Yonezawa, Y, Ishibashi, M, Matsumoto, F, Tamada, T, Tokunaga, H, Tokunaga, M, Kuroki, R. | | Deposit date: | 2013-05-17 | | Release date: | 2014-03-12 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural characteristics of alkaline phosphatase from the moderately halophilic bacterium Halomonas sp. 593.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
3VUG
 
 | | Crystal structure of a cysteine-deficient mutant M2 in MAP kinase JNK1 | | Descriptor: | Mitogen-activated protein kinase 8, Peptide from C-Jun-amino-terminal kinase-interacting protein 1, SULFATE ION | | Authors: | Nakaniwa, T, Kinoshita, T, Inoue, T. | | Deposit date: | 2012-06-28 | | Release date: | 2013-02-13 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (3.24 Å) | | Cite: | Seven cysteine-deficient mutants depict the interplay between thermal and chemical stabilities of individual cysteine residues in mitogen-activated protein kinase c-Jun N-terminal kinase 1
Biochemistry, 51, 2012
|
|
3VUD
 
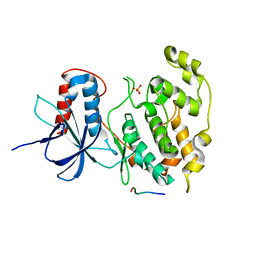 | | Crystal structure of a cysteine-deficient mutant M1 in MAP kinase JNK1 | | Descriptor: | Mitogen-activated protein kinase 8, Peptide from C-Jun-amino-terminal kinase-interacting protein 1, SULFATE ION | | Authors: | Nakaniwa, T, Kinoshita, T, Inoue, T. | | Deposit date: | 2012-06-28 | | Release date: | 2013-02-13 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Seven cysteine-deficient mutants depict the interplay between thermal and chemical stabilities of individual cysteine residues in mitogen-activated protein kinase c-Jun N-terminal kinase 1
Biochemistry, 51, 2012
|
|
