5Z0R
 
 | | Structural insight into the Zika virus capsid encapsulating the viral genome | | Descriptor: | Extracellular solute-binding protein family 1,viral genome protein, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Li, T, Zhao, Q, Yang, X, Chen, C, Yang, K, Wu, C, Zhang, T, Duan, Y, Xue, X, Mi, K, Ji, X, Wang, Z, Yang, H. | | Deposit date: | 2017-12-20 | | Release date: | 2018-04-11 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structural insight into the Zika virus capsid encapsulating the viral genome.
Cell Res., 28, 2018
|
|
5X8Y
 
 | | A Mutation identified in Neonatal Microcephaly Destabilizes Zika Virus NS1 Assembly in vitro | | Descriptor: | ZIKV NS1 | | Authors: | Wang, D, Chen, C, Liu, S, Zhou, H, Yang, K, Zhao, Q, Ji, X, Chen, C, Xie, W, Wang, Z, Mi, L.Z, Yang, H. | | Deposit date: | 2017-03-03 | | Release date: | 2017-05-17 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.817 Å) | | Cite: | A Mutation Identified in Neonatal Microcephaly Destabilizes Zika Virus NS1 Assembly in Vitro
Sci Rep, 7, 2017
|
|
3V3Q
 
 | | Crystal Structure of Human Nur77 Ligand-binding Domain in Complex with Ethyl 2-[2,3,4 trimethoxy-6(1-octanoyl)phenyl]acetate | | Descriptor: | GLYCEROL, Nuclear receptor subfamily 4 group A member 1, SODIUM ION, ... | | Authors: | Zhang, Q, Shi, C, Yang, K, Chen, Y, Zhan, Y, Wu, Q, Lin, T. | | Deposit date: | 2011-12-14 | | Release date: | 2012-09-26 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | The orphan nuclear receptor Nur77 regulates LKB1 localization and activates AMPK
Nat.Chem.Biol., 8, 2012
|
|
3V3E
 
 | | Crystal Structure of the Human Nur77 Ligand-binding Domain | | Descriptor: | GLYCEROL, Nuclear receptor subfamily 4 group A member 1 | | Authors: | Zhang, Q, Shi, C, Yang, K, Chen, Y, Zhan, Y, Wu, Q, Lin, T. | | Deposit date: | 2011-12-13 | | Release date: | 2012-09-26 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | The orphan nuclear receptor Nur77 regulates LKB1 localization and activates AMPK
Nat.Chem.Biol., 8, 2012
|
|
2HBX
 
 | | Crystal Structure of alpha-Amino-beta-Carboxymuconate-epsilon-Semialdehyde-Decarboxylase (ACMSD) | | Descriptor: | 2-amino-3-carboxymuconate 6-semialdehyde decarboxylase, COBALT (II) ION | | Authors: | Martynowski, D, Eyobo, Y, Li, T, Yang, K, Liu, A, Zhang, H. | | Deposit date: | 2006-06-14 | | Release date: | 2006-09-19 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal Structure of alpha-Amino-beta-carboxymuconate-epsilon-semialdehyde Decarboxylase: Insight into the Active Site and Catalytic Mechanism of a Novel Decarboxylation Reaction.
Biochemistry, 45, 2006
|
|
2HBV
 
 | | Crystal Structure of alpha-Amino-beta-Carboxymuconate-epsilon-Semialdehyde-Decarboxylase (ACMSD) | | Descriptor: | 2-amino-3-carboxymuconate 6-semialdehyde decarboxylase, MAGNESIUM ION, ZINC ION | | Authors: | Martynowski, D, Eyobo, Y, Li, T, Yang, K, Liu, A, Zhang, H. | | Deposit date: | 2006-06-14 | | Release date: | 2006-09-19 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystal Structure of alpha-Amino-beta-carboxymuconate-epsilon-semialdehyde Decarboxylase: Insight into the Active Site and Catalytic Mechanism of a Novel Decarboxylation Reaction.
Biochemistry, 45, 2006
|
|
3AKA
 
 | | Structural basis for prokaryotic calcium-mediated regulation by a Streptomyces coelicolor calcium-binding protein | | Descriptor: | CALCIUM ION, Putative calcium binding protein | | Authors: | Zhao, X, Pang, H, Wang, S, Zhou, W, Yang, K, Bartlam, M. | | Deposit date: | 2010-07-09 | | Release date: | 2011-01-26 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural basis for prokaryotic calciummediated regulation by a Streptomyces coelicolor calcium binding protein
Protein Cell, 1, 2010
|
|
4KJK
 
 | | Room Temperature WT DHFR | | Descriptor: | CALCIUM ION, Dihydrofolate reductase, FOLIC ACID, ... | | Authors: | van den Bedem, H, Bhabha, G, Yang, K, Wright, P.E, Fraser, J.S. | | Deposit date: | 2013-05-03 | | Release date: | 2013-08-21 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.351 Å) | | Cite: | Automated identification of functional dynamic contact networks from X-ray crystallography.
Nat.Methods, 10, 2013
|
|
4KJJ
 
 | | Cryogenic WT DHFR | | Descriptor: | Dihydrofolate reductase, FOLIC ACID, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | van den Bedem, H, Bhabha, G, Yang, K, Wright, P.E, Fraser, J.S. | | Deposit date: | 2013-05-03 | | Release date: | 2013-08-21 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Automated identification of functional dynamic contact networks from X-ray crystallography.
Nat.Methods, 10, 2013
|
|
4KJL
 
 | | Room Temperature N23PPS148A DHFR | | Descriptor: | Dihydrofolate reductase, FOLIC ACID, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | van den Bedem, H, Bhabha, G, Yang, K, Wright, P.E, Fraser, J.S. | | Deposit date: | 2013-05-03 | | Release date: | 2013-08-21 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.38 Å) | | Cite: | Automated identification of functional dynamic contact networks from X-ray crystallography.
Nat.Methods, 10, 2013
|
|
4ICY
 
 | | Tracing the Evolution of Angucyclinone Monooxygenases: Structural Determinants for C-12b Hydroxylation and Substrate Inhibition in PgaE | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Kallio, P, Patrikainen, P, Belogurov, G, Mantsala, P, Yang, K, Niemi, J, Metsa-Ketela, M. | | Deposit date: | 2012-12-11 | | Release date: | 2013-06-12 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Tracing the Evolution of Angucyclinone Monooxygenases: Structural Determinants for C-12b Hydroxylation and Substrate Inhibition in PgaE.
Biochemistry, 52, 2013
|
|
7RVO
 
 | | Structure of the SARS-CoV-2 main protease in complex with inhibitor MPI13 | | Descriptor: | 3C-like proteinase, N-[(benzyloxy)carbonyl]-L-valyl-3-cyclopropyl-N-{(2S)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-L-alaninamide | | Authors: | Yang, K, Sankaran, B, Liu, W. | | Deposit date: | 2021-08-19 | | Release date: | 2022-07-20 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | A multi-pronged evaluation of aldehyde-based tripeptidyl main protease inhibitors as SARS-CoV-2 antivirals.
Eur.J.Med.Chem., 240, 2022
|
|
7RVR
 
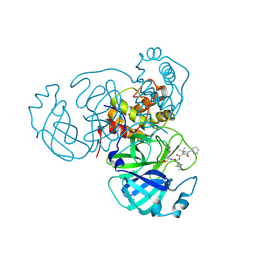 | | Structure of the SARS-CoV-2 main protease in complex with inhibitor MPI18 | | Descriptor: | 3C-like proteinase, N-[(benzyloxy)carbonyl]-3-methyl-L-valyl-N-{(2S)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-4-methyl-L-leucinamide | | Authors: | Yang, K, Liu, W. | | Deposit date: | 2021-08-19 | | Release date: | 2022-07-20 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.46 Å) | | Cite: | A multi-pronged evaluation of aldehyde-based tripeptidyl main protease inhibitors as SARS-CoV-2 antivirals.
Eur.J.Med.Chem., 240, 2022
|
|
7RVS
 
 | | Structure of the SARS-CoV-2 main protease in complex with inhibitor MPI19 | | Descriptor: | 3C-like proteinase, N-[(benzyloxy)carbonyl]-3-methyl-L-valyl-3-cyclopropyl-N-{(2S)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-L-alaninamide | | Authors: | Yang, K, Liu, W. | | Deposit date: | 2021-08-19 | | Release date: | 2022-07-20 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | A multi-pronged evaluation of aldehyde-based tripeptidyl main protease inhibitors as SARS-CoV-2 antivirals.
Eur.J.Med.Chem., 240, 2022
|
|
7RW0
 
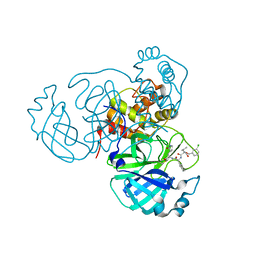 | | Structure of the SARS-CoV-2 main protease in complex with inhibitor MPI27 | | Descriptor: | 3C-like proteinase, N-{[(3-chlorophenyl)methoxy]carbonyl}-L-valyl-3-cyclohexyl-N-{(2S)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-L-alaninamide | | Authors: | Yang, K, Liu, W. | | Deposit date: | 2021-08-19 | | Release date: | 2022-07-20 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | A multi-pronged evaluation of aldehyde-based tripeptidyl main protease inhibitors as SARS-CoV-2 antivirals.
Eur.J.Med.Chem., 240, 2022
|
|
7RVM
 
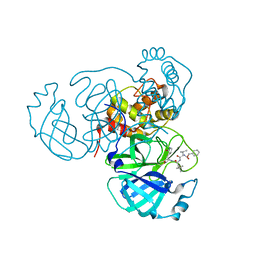 | | Structure of the SARS-CoV-2 main protease in complex with inhibitor MPI11 | | Descriptor: | 3C-like proteinase, N-[(benzyloxy)carbonyl]-L-valyl-N-{(2S)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-4-methyl-L-leucinamide | | Authors: | Yang, K, Liu, W. | | Deposit date: | 2021-08-19 | | Release date: | 2022-07-20 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | A multi-pronged evaluation of aldehyde-based tripeptidyl main protease inhibitors as SARS-CoV-2 antivirals.
Eur.J.Med.Chem., 240, 2022
|
|
7RVU
 
 | | Structure of the SARS-CoV-2 main protease in complex with inhibitor MPI21 | | Descriptor: | 3C-like proteinase, N-[(benzyloxy)carbonyl]-3-methyl-L-isovalyl-N-{(2S)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-4-methyl-L-leucinamide | | Authors: | Yang, K, Sankaran, B, Liu, W. | | Deposit date: | 2021-08-19 | | Release date: | 2022-07-20 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | A multi-pronged evaluation of aldehyde-based tripeptidyl main protease inhibitors as SARS-CoV-2 antivirals.
Eur.J.Med.Chem., 240, 2022
|
|
7RVW
 
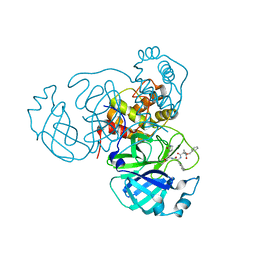 | | Structure of the SARS-CoV-2 main protease in complex with inhibitor MPI23 | | Descriptor: | 3C-like proteinase, benzyl (1-{[(2S)-3-cyclohexyl-1-({(2S)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}amino)-1-oxopropan-2-yl]carbamoyl}cyclopropyl)carbamate | | Authors: | Yang, K, Liu, W. | | Deposit date: | 2021-08-19 | | Release date: | 2022-07-20 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | A multi-pronged evaluation of aldehyde-based tripeptidyl main protease inhibitors as SARS-CoV-2 antivirals.
Eur.J.Med.Chem., 240, 2022
|
|
7RW1
 
 | |
7RVV
 
 | | Structure of the SARS-CoV-2 main protease in complex with inhibitor MPI22 | | Descriptor: | 3C-like proteinase, N-[(benzyloxy)carbonyl]-2-methyl-L-alanyl-N-{(2S)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-4-methyl-L-leucinamide | | Authors: | Yang, K, Liu, W. | | Deposit date: | 2021-08-19 | | Release date: | 2022-07-20 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | A multi-pronged evaluation of aldehyde-based tripeptidyl main protease inhibitors as SARS-CoV-2 antivirals.
Eur.J.Med.Chem., 240, 2022
|
|
7RVZ
 
 | | Structure of the SARS-CoV-2 main protease in complex with inhibitor MPI26 | | Descriptor: | 3C-like proteinase, O-tert-butyl-N-{[(3-chlorophenyl)methoxy]carbonyl}-L-threonyl-N-{(2S)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-L-leucinamide | | Authors: | Yang, K, Sankaran, B, Liu, W. | | Deposit date: | 2021-08-19 | | Release date: | 2022-07-20 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | A multi-pronged evaluation of aldehyde-based tripeptidyl main protease inhibitors as SARS-CoV-2 antivirals.
Eur.J.Med.Chem., 240, 2022
|
|
7RVQ
 
 | | Structure of the SARS-CoV-2 main protease in complex with inhibitor MPI16 | | Descriptor: | 3C-like proteinase, N-[(benzyloxy)carbonyl]-O-tert-butyl-L-threonyl-N-{(2S)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-4-methyl-L-leucinamide | | Authors: | Yang, K, Liu, W. | | Deposit date: | 2021-08-19 | | Release date: | 2022-07-20 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | A multi-pronged evaluation of aldehyde-based tripeptidyl main protease inhibitors as SARS-CoV-2 antivirals.
Eur.J.Med.Chem., 240, 2022
|
|
7RVX
 
 | | Structure of the SARS-CoV-2 main protease in complex with inhibitor MPI24 | | Descriptor: | 3C-like proteinase, benzyl [(1S)-1-cyclopropyl-2-{[(2S)-3-cyclopropyl-1-({(2S)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}amino)-1-oxopropan-2-yl]amino}-2-oxoethyl]carbamate | | Authors: | Yang, K, Liu, W. | | Deposit date: | 2021-08-19 | | Release date: | 2022-07-20 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | A multi-pronged evaluation of aldehyde-based tripeptidyl main protease inhibitors as SARS-CoV-2 antivirals.
Eur.J.Med.Chem., 240, 2022
|
|
7RVP
 
 | | Structure of the SARS-CoV-2 main protease in complex with inhibitor MPI14 | | Descriptor: | 3C-like proteinase, N-[(benzyloxy)carbonyl]-L-valyl-3-furan-2-yl-N-{(2S)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-L-alaninamide | | Authors: | Yang, K, Sankaran, B, Liu, W. | | Deposit date: | 2021-08-19 | | Release date: | 2022-07-20 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | A multi-pronged evaluation of aldehyde-based tripeptidyl main protease inhibitors as SARS-CoV-2 antivirals.
Eur.J.Med.Chem., 240, 2022
|
|
7RVT
 
 | | Structure of the SARS-CoV-2 main protease in complex with inhibitor MPI20 | | Descriptor: | 3C-like proteinase, N~2~-[(2S)-2-{[(benzyloxy)carbonyl]amino}-2-cyclopropylacetyl]-N-{(2S)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-4-methyl-L-leucinamide | | Authors: | Yang, K, Sankaran, B, Liu, W. | | Deposit date: | 2021-08-19 | | Release date: | 2022-07-20 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | A multi-pronged evaluation of aldehyde-based tripeptidyl main protease inhibitors as SARS-CoV-2 antivirals.
Eur.J.Med.Chem., 240, 2022
|
|
