2V3X
 
 | |
2V3Y
 
 | |
1PV9
 
 | | Prolidase from Pyrococcus furiosus | | Descriptor: | Xaa-Pro dipeptidase, ZINC ION | | Authors: | Maher, M.J, Ghosh, M, Grunden, A.M, Menon, A.L, Adams, M.W, Freeman, H.C, Guss, J.M. | | Deposit date: | 2003-06-27 | | Release date: | 2004-03-23 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of the Prolidase from Pyrococcus furiosus.
Biochemistry, 43, 2004
|
|
1KSI
 
 | | CRYSTAL STRUCTURE OF A EUKARYOTIC (PEA SEEDLING) COPPER-CONTAINING AMINE OXIDASE AT 2.2A RESOLUTION | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, COPPER (II) ION, ... | | Authors: | Wilce, M.C.J, Kumar, V, Freeman, H.C, Guss, J.M. | | Deposit date: | 1996-07-20 | | Release date: | 1997-12-24 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of a eukaryotic (pea seedling) copper-containing amine oxidase at 2.2 A resolution.
Structure, 4, 1996
|
|
1KCI
 
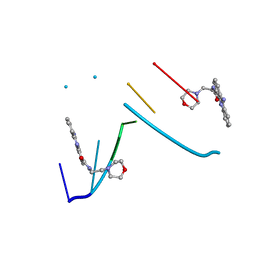 | | Crystal Structure of 9-amino-N-[2-(4-morpholinyl)ethyl]-4-acridinecarboxamide Bound to d(CGTACG)2 | | Descriptor: | 5'-D(*CP*GP*TP*AP*CP*G)-3', 9-AMINO-N-[2-(4-MORPHOLINYL)ETHYL]-4-ACRIDINECARBOXAMIDE | | Authors: | Adams, A, Guss, J.M, Denny, W.A, Wakelin, L.P.G. | | Deposit date: | 2001-11-08 | | Release date: | 2002-02-01 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of 9-amino-N-[2-(4-morpholinyl)ethyl]-4-acridinecarboxamide bound to d(CGTACG)2: implications for structure-activity relationships of acridinecarboxamide topoisomerase poisons.
Nucleic Acids Res., 30, 2002
|
|
1JAW
 
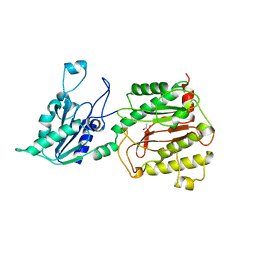 | | AMINOPEPTIDASE P FROM E. COLI LOW PH FORM | | Descriptor: | ACETATE ION, AMINOPEPTIDASE P, MANGANESE (II) ION | | Authors: | Wilce, M.C.J, Bond, C.S, Lilley, P.E, Dixon, N.E, Freeman, H.C, Guss, J.M. | | Deposit date: | 1997-12-22 | | Release date: | 1999-04-06 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structure and mechanism of a proline-specific aminopeptidase from Escherichia coli.
Proc.Natl.Acad.Sci.USA, 95, 1998
|
|
3SZK
 
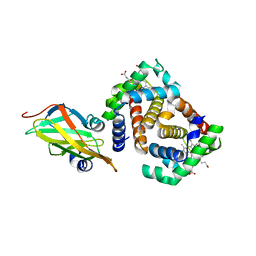 | | Crystal Structure of Human metHaemoglobin Complexed with the First NEAT Domain of IsdH from Staphylococcus aureus | | Descriptor: | Hemoglobin subunit alpha, Hemoglobin subunit beta, Iron-regulated surface determinant protein H, ... | | Authors: | Jacques, D.A, Kumar, K.K, Guss, J.M, Gell, D.A. | | Deposit date: | 2011-07-19 | | Release date: | 2011-09-14 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.01 Å) | | Cite: | Structural basis for hemoglobin capture by Staphylococcus aureus cell-surface protein, IsdH
J.Biol.Chem., 286, 2011
|
|
3S48
 
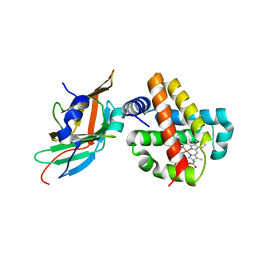 | | Human Alpha-Haemoglobin Complexed with the First NEAT Domain of IsdH from Staphylococcus aureus | | Descriptor: | Hemoglobin subunit alpha, Iron-regulated surface determinant protein H, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Kumar, K.K, Jacques, D.A, Caradoc-Davies, T.T, Guss, J.M, Gell, D.A. | | Deposit date: | 2011-05-19 | | Release date: | 2012-05-23 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.05 Å) | | Cite: | The structure of alpha-haemoglobin in complex with a haemoglobin-binding domain from Staphylococcus aureus reveals the elusive alpha-haemoglobin dimerization interface
ACTA CRYSTALLOGR.,SECT.F, 70, 2014
|
|
3TAH
 
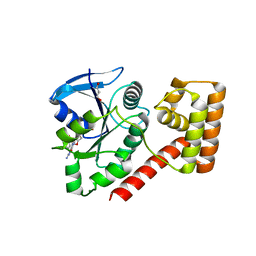 | | Crystal structure of an S. thermophilus NFeoB N11A mutant bound to mGDP | | Descriptor: | 3'-O-(N-methylanthraniloyl)guanosine-5'-diphosphate, Ferrous iron uptake transporter protein B, GLYCEROL, ... | | Authors: | Ash, M.R, Maher, M.J, Guss, J.M, Jormakka, M. | | Deposit date: | 2011-08-04 | | Release date: | 2011-12-14 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | The structure of an N11A mutant of the G-protein domain of FeoB
Acta Crystallogr.,Sect.F, 67, 2011
|
|
3UK3
 
 | | Crystal structure of ZNF217 bound to DNA | | Descriptor: | 5'-D(*AP*AP*TP*GP*CP*AP*GP*AP*AP*TP*CP*GP*AP*TP*TP*CP*TP*GP*CP*A)-3', 5'-D(*TP*TP*TP*GP*CP*AP*GP*AP*AP*TP*CP*GP*AP*TP*TP*CP*TP*GP*CP*A)-3', CHLORIDE ION, ... | | Authors: | Vandevenne, M.S, Jacques, D.A, Guss, J.M, Mackay, J.P. | | Deposit date: | 2011-11-08 | | Release date: | 2012-12-05 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Rediscovering DNA recognition by classical Zinc Fingers.
To be Published
|
|
3TU6
 
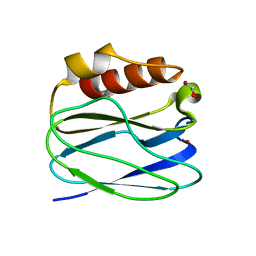 | | The Structure of a Pseudoazurin From Sinorhizobium meliltoi | | Descriptor: | COPPER (II) ION, GLYCEROL, Pseudoazurin (Blue copper protein) | | Authors: | Laming, E.M, McGrath, A.P, Guss, J.M, Maher, M.J. | | Deposit date: | 2011-09-16 | | Release date: | 2012-05-30 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The X-ray crystal structure of a pseudoazurin from Sinorhizobium meliloti.
J.Inorg.Biochem., 115, 2012
|
|
3K5T
 
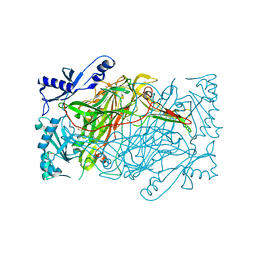 | | Crystal structure of human diamine oxidase in space group C2221 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | McGrath, A.P, Guss, J.M. | | Deposit date: | 2009-10-08 | | Release date: | 2010-02-09 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | A new crystal form of human diamine oxidase.
Acta Crystallogr.,Sect.F, 66, 2010
|
|
3SS8
 
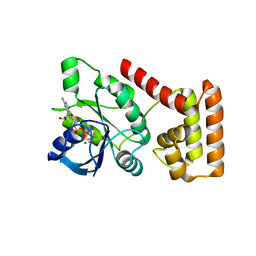 | | Crystal structure of NFeoB from S. thermophilus bound to GDP.AlF4- and K+ | | Descriptor: | Ferrous iron uptake transporter protein B, GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Ash, M.R, Maher, M.J, Guss, J.M, Jormakka, M. | | Deposit date: | 2011-07-08 | | Release date: | 2011-08-24 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | The initiation of GTP hydrolysis by the G-domain of FeoB: insights from a transition-state complex structure
Plos One, 6, 2011
|
|
3HI7
 
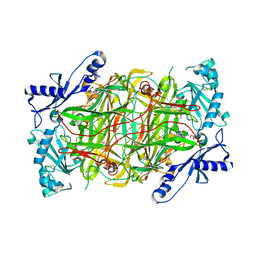 | | Crystal structure of human diamine oxidase | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Amiloride-sensitive amine oxidase, CALCIUM ION, ... | | Authors: | McGrath, A.P, Guss, J.M. | | Deposit date: | 2009-05-19 | | Release date: | 2009-10-20 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.799 Å) | | Cite: | Structure and inhibition of human diamine oxidase
Biochemistry, 48, 2009
|
|
3HIG
 
 | |
3HII
 
 | |
1DL8
 
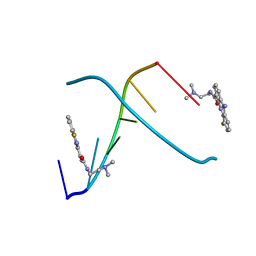 | | CRYSTAL STRUCTURE OF 5-F-9-AMINO-(N-(2-DIMETHYLAMINO)ETHYL)ACRIDINE-4-CARBOXAMIDE BOUND TO D(CGTACG)2 | | Descriptor: | 5-FLUORO-9-AMINO-(N-(2-DIMETHYLAMINO)ETHYL)ACRIDINE-4-CARBOXAMIDE, DNA (5'-D(*CP*GP*TP*AP*CP*G)-3') | | Authors: | Adams, A, Guss, J.M, Collyer, C.A, Denny, W.A, Wakelin, L.P. | | Deposit date: | 1999-12-08 | | Release date: | 2000-10-30 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Acridinecarboxamide topoisomerase poisons: structural and kinetic studies of the DNA complexes of 5-substituted 9-amino-(N-(2-dimethylamino)ethyl)acridine-4-carboxamides.
Mol.Pharmacol., 58, 2000
|
|
1FN2
 
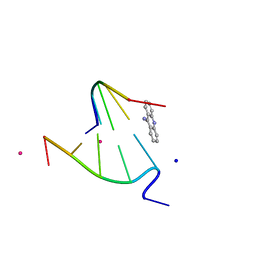 | | 9-AMINO-(N-(2-DIMETHYLAMINO)BUTYL)ACRIDINE-4-CARBOXAMIDE BOUND TO D(CGTACG)2 | | Descriptor: | 9-AMINO-(N-(2-DIMETHYLAMINO)BUTYL)ACRIDINE-4-CARBOXAMIDE, COBALT (II) ION, DNA (5'-D(*CP*GP*TP*AP*CP*G)-3'), ... | | Authors: | Adams, A, Guss, J.M, Collyer, C.A, Denny, W.A, Wakelin, L.P.G. | | Deposit date: | 2000-08-19 | | Release date: | 2000-10-30 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | A novel form of intercalation involving four DNA duplexes in an acridine-4-carboxamide complex of d(CGTACG)(2).
Nucleic Acids Res., 28, 2000
|
|
1FN1
 
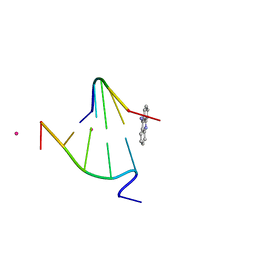 | | CRYSTAL STRUCTURE OF 9-AMINO-(N-(2-DIMETHYLAMINO)BUTYL)ACRIDINE-4-CARBOXAMIDE BOUND TO D(CG(5BR)UACG)2 | | Descriptor: | 9-AMINO-(N-(2-DIMETHYLAMINO)BUTYL)ACRIDINE-4-CARBOXAMIDE, COBALT (II) ION, DNA (5'-D(*CP*GP*(BRO)UP*AP*CP*G)-3'), ... | | Authors: | Adams, A, Guss, J.M, Collyer, C.A, Denny, W.A, Wakelin, L.P.G. | | Deposit date: | 2000-08-19 | | Release date: | 2000-10-30 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | A novel form of intercalation involving four DNA duplexes in an acridine-4-carboxamide complex of d(CGTACG)(2).
Nucleic Acids Res., 28, 2000
|
|
1RQY
 
 | | 9-amino-[N-(2-dimethylamino)proply]-acridine-4-carboxamide bound to d(CGTACG)2 | | Descriptor: | 5'-D(CP*GP*TP*AP*CP*G)-3', 9-AMINO-N-[3-(DIMETHYLAMINO)PROPYL]ACRIDINE-4-CARBOXAMIDE, COBALT (II) ION, ... | | Authors: | Adams, A, Guss, J.M, Denny, W.A, Wakelin, L.P. | | Deposit date: | 2003-12-07 | | Release date: | 2004-05-11 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structure of 9-amino-[N-(2-dimethylamino)propyl]acridine-4-carboxamide bound to d(CGTACG)(2): a comparison of structures of d(CGTACG)(2) complexed with intercalatorsin the presence of cobalt.
Acta Crystallogr.,Sect.D, 60, 2004
|
|
3OPF
 
 | | Crystal structure of TTHA0988 in space group P212121 | | Descriptor: | GLYCEROL, Putative uncharacterized protein TTHA0988, SULFATE ION | | Authors: | Jacques, D.A, Kuramitsu, S, Yokoyama, S, Trewhella, J, Guss, J.M, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2010-09-01 | | Release date: | 2011-02-02 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | The structure of TTHA0988 from Thermus thermophilus, a KipI-KipA homologue incorrectly annotated as an allophanate hydrolase
Acta Crystallogr.,Sect.D, 67, 2011
|
|
3OVU
 
 | | Crystal Structure of Human Alpha-Haemoglobin Complexed with AHSP and the First NEAT Domain of IsdH from Staphylococcus aureus | | Descriptor: | Alpha-hemoglobin-stabilizing protein, Hemoglobin subunit alpha, Iron-regulated surface determinant protein H, ... | | Authors: | Jacques, D.A, Krishna Kumar, K, Caradoc-Davies, T.T, Langley, D.B, Mackay, J.P, Guss, J.M, Gell, D.A. | | Deposit date: | 2010-09-17 | | Release date: | 2011-09-21 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.83 Å) | | Cite: | A new haem pocket structure in alpha-haemoglobin
To be Published
|
|
3ORE
 
 | | Crystal structure of TTHA0988 in space group P6522 | | Descriptor: | Putative uncharacterized protein TTHA0988 | | Authors: | Jacques, D.A, Kuramitsu, S, Yokoyama, S, Trewhella, J, Guss, J.M, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2010-09-07 | | Release date: | 2011-02-02 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | The structure of TTHA0988 from Thermus thermophilus, a KipI-KipA homologue incorrectly annotated as an allophanate hydrolase
Acta Crystallogr.,Sect.D, 67, 2011
|
|
1B2O
 
 | | CLOSTRIDIUM PASTEURIANUM RUBREDOXIN G10VG43A MUTANT | | Descriptor: | FE (III) ION, PROTEIN (RUBREDOXIN) | | Authors: | Maher, M.J, Guss, J.M, Wilce, M.C.J, Wedd, A.G. | | Deposit date: | 1998-11-30 | | Release date: | 1999-05-27 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Rubredoxin from Clostridium pasteurianum. Structures of G10A, G43A and G10VG43A mutant proteins. Mutation of conserved glycine 10 to valine causes the 9-10 peptide link to invert.
Acta Crystallogr.,Sect.D, 55, 1999
|
|
1B2J
 
 | | CLOSTRIDIUM PASTEURIANUM RUBREDOXIN G43A MUTANT | | Descriptor: | FE (III) ION, PROTEIN (RUBREDOXIN) | | Authors: | Maher, M.J, Guss, J.M, Wilce, M.C.J, Wedd, A.G. | | Deposit date: | 1998-11-27 | | Release date: | 1999-05-27 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Rubredoxin from Clostridium pasteurianum. Structures of G10A, G43A and G10VG43A mutant proteins. Mutation of conserved glycine 10 to valine causes the 9-10 peptide link to invert.
Acta Crystallogr.,Sect.D, 55, 1999
|
|
