7F0I
 
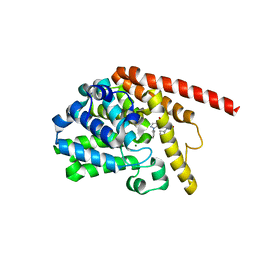 | | phosphodiesterase-9A in complex with inhibitor 4b | | Descriptor: | 1-cyclopentyl-6-[[(2R)-1-(6-fluoranyl-2-azaspiro[3.3]heptan-2-yl)-1-oxidanylidene-propan-2-yl]amino]-5H-pyrazolo[3,4-d]pyrimidin-4-one, Isoform PDE9A2 of High affinity cGMP-specific 3',5'-cyclic phosphodiesterase 9A, MAGNESIUM ION, ... | | Authors: | Wu, Y, Huang, Y.Y, Luo, H.B. | | Deposit date: | 2021-06-04 | | Release date: | 2022-04-13 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.70000887 Å) | | Cite: | Discovery of Potent Phosphodiesterase-9 Inhibitors for the Treatment of Hepatic Fibrosis
J.Med.Chem., 64, 2021
|
|
7F2B
 
 | |
7F2E
 
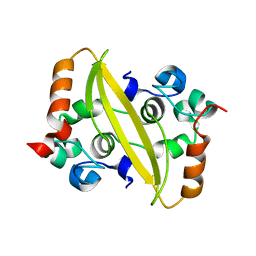 | | SARS-CoV-2 nucleocapsid protein C-terminal domain (dodecamer) | | Descriptor: | Nucleoprotein, PHOSPHATE ION | | Authors: | Liu, C, Jiang, H. | | Deposit date: | 2021-06-10 | | Release date: | 2021-10-20 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Crystal structures of the SARS-CoV-2 nucleocapsid protein C-terminal domain and development of nucleocapsid-targeting nanobodies.
Febs J., 289, 2022
|
|
7TXW
 
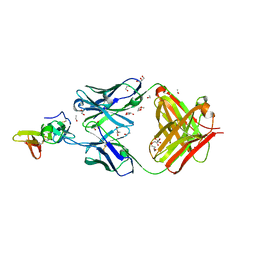 | | Crystal structure of the complex of the malaria sexual stage protein and vaccine target Pfs25 with the Fab fragment of a transmission blocking antibody 1G2 | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, ETHANOL, ... | | Authors: | Singh, K, Gittis, A.G, Garboczi, D.N. | | Deposit date: | 2022-02-10 | | Release date: | 2023-04-12 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | Structural and immunological differences in Plasmodium falciparum sexual stage transmission-blocking vaccines comprised of Pfs25-EPA nanoparticles.
Npj Vaccines, 8, 2023
|
|
7D6N
 
 | |
7V61
 
 | | ACE2 -Targeting Monoclonal Antibody as Potent and Broad-Spectrum Coronavirus Blocker | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 3E8, ... | | Authors: | Yan, R.H, Zhang, Y.Y, Li, Y.N, Zhou, Q. | | Deposit date: | 2021-08-18 | | Release date: | 2022-06-22 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | ACE2-targeting monoclonal antibody as potent and broad-spectrum coronavirus blocker.
Signal Transduct Target Ther, 6, 2021
|
|
7X08
 
 | | S protein of SARS-CoV-2 in complex with 2G1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, LINOLEIC ACID, ... | | Authors: | Guo, Y.Y, Zhang, Y.Y, Zhou, Q. | | Deposit date: | 2022-02-21 | | Release date: | 2022-03-09 | | Last modified: | 2023-11-15 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Broad ultra-potent neutralization of SARS-CoV-2 variants by monoclonal antibodies specific to the tip of RBD.
Cell Discov, 8, 2022
|
|
7VAH
 
 | |
8GTZ
 
 | |
7TY7
 
 | | Cryo-EM structure of human Anion Exchanger 1 bound to Bicarbonate | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, BICARBONATE ION, ... | | Authors: | Capper, M.J, Mathiharan, Y.K, Yang, S, Stone, A.C, Wacker, D. | | Deposit date: | 2022-02-11 | | Release date: | 2023-08-16 | | Last modified: | 2023-11-01 | | Method: | ELECTRON MICROSCOPY (3.37 Å) | | Cite: | Substrate binding and inhibition of the anion exchanger 1 transporter.
Nat.Struct.Mol.Biol., 30, 2023
|
|
7TY6
 
 | | Cryo-EM structure of human Anion Exchanger 1 bound to 4,4'-Diisothiocyanatodihydrostilbene-2,2'-Disulfonic Acid (H2DIDS) | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 4,4'-Diisothiocyano-2,2'-stilbenedisulfonic acid, ... | | Authors: | Capper, M.J, Mathiharan, Y.K, Yang, S, Stone, A.C, Wacker, D. | | Deposit date: | 2022-02-11 | | Release date: | 2023-08-16 | | Last modified: | 2023-11-01 | | Method: | ELECTRON MICROSCOPY (2.98 Å) | | Cite: | Substrate binding and inhibition of the anion exchanger 1 transporter.
Nat.Struct.Mol.Biol., 30, 2023
|
|
7TY4
 
 | | Cryo-EM structure of human Anion Exchanger 1 | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Band 3 anion transport protein, ... | | Authors: | Capper, M.J, Mathiharan, Y.K, Yang, S, Stone, A.C, Wacker, D. | | Deposit date: | 2022-02-11 | | Release date: | 2023-08-16 | | Last modified: | 2023-11-01 | | Method: | ELECTRON MICROSCOPY (2.99 Å) | | Cite: | Substrate binding and inhibition of the anion exchanger 1 transporter.
Nat.Struct.Mol.Biol., 30, 2023
|
|
7TYA
 
 | | Cryo-EM structure of human Anion Exchanger 1 modified with Diethyl Pyrocarbonate (DEPC) | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Band 3 anion transport protein, ... | | Authors: | Capper, M.J, Mathiharan, Y.K, Yang, S, Stone, A.C, Wacker, D. | | Deposit date: | 2022-02-11 | | Release date: | 2023-08-16 | | Last modified: | 2023-11-01 | | Method: | ELECTRON MICROSCOPY (3.07 Å) | | Cite: | Substrate binding and inhibition of the anion exchanger 1 transporter.
Nat.Struct.Mol.Biol., 30, 2023
|
|
7TY8
 
 | | Cryo-EM structure of human Anion Exchanger 1 bound to Niflumic Acid | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-{[3-(TRIFLUOROMETHYL)PHENYL]AMINO}NICOTINIC ACID, ... | | Authors: | Capper, M.J, Mathiharan, Y.K, Yang, S, Stone, A.C, Wacker, D. | | Deposit date: | 2022-02-11 | | Release date: | 2023-08-16 | | Last modified: | 2023-11-01 | | Method: | ELECTRON MICROSCOPY (3.18 Å) | | Cite: | Substrate binding and inhibition of the anion exchanger 1 transporter.
Nat.Struct.Mol.Biol., 30, 2023
|
|
