7CI1
 
 | | Crystal structure of AcrVA2 | | Descriptor: | 1,2-ETHANEDIOL, AcrVA2, SPERMIDINE | | Authors: | Chen, P, Cheng, Z, Wang, Y. | | Deposit date: | 2020-07-07 | | Release date: | 2020-10-14 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural Study on Anti-CRISPR Protein AcrVA2
Prog.Biochem.Biophys., 2021
|
|
6IDZ
 
 | | Crystal structure of H7 hemagglutinin mutant H7-SVTQ ( A138S, P221T, L226Q) with 3'SLN | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Hemagglutinin HA1 chain, Hemagglutinin HA2 chain, ... | | Authors: | Gao, G.F, Xu, Y, Qi, J.X. | | Deposit date: | 2018-09-12 | | Release date: | 2019-11-27 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.707 Å) | | Cite: | Avian-to-Human Receptor-Binding Adaptation of Avian H7N9 Influenza Virus Hemagglutinin.
Cell Rep, 29, 2019
|
|
7CI2
 
 | |
6IDA
 
 | | Crystal structure of H7 hemagglutinin mutant H7-SVTQ ( A138S, P221T, L226Q) from the influenza virus A/Anhui/1/2013 (H7N9) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Hemagglutinin HA1 chain, Hemagglutinin HA2 chain | | Authors: | Gao, G.F, Xu, Y, Qi, J.X. | | Deposit date: | 2018-09-09 | | Release date: | 2019-11-27 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (3.104 Å) | | Cite: | Avian-to-Human Receptor-Binding Adaptation of Avian H7N9 Influenza Virus Hemagglutinin.
Cell Rep, 29, 2019
|
|
6ID8
 
 | | Crystal structure of H7 hemagglutinin mutant H7-SVTL ( A138S, P221T) from the influenza virus A/Anhui/1/2013 (H7N9) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Hemagglutinin HA1 chain, Hemagglutinin HA2 chain | | Authors: | Gao, G.F, Xu, Y, Qi, J.X. | | Deposit date: | 2018-09-09 | | Release date: | 2019-11-27 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.902 Å) | | Cite: | Avian-to-Human Receptor-Binding Adaptation of Avian H7N9 Influenza Virus Hemagglutinin.
Cell Rep, 29, 2019
|
|
6ID3
 
 | | Crystal structure of H7 hemagglutinin mutant H7-SGPL ( A138S, V186G) from the influenza virus A/Anhui/1/2013 (H7N9) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Hemagglutinin HA1 chain, Hemagglutinin HA2 chain | | Authors: | Gao, G.F, Xu, Y, Qi, J.X. | | Deposit date: | 2018-09-08 | | Release date: | 2019-11-27 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Avian-to-Human Receptor-Binding Adaptation of Avian H7N9 Influenza Virus Hemagglutinin.
Cell Rep, 29, 2019
|
|
6IDD
 
 | | Crystal structure of H7 hemagglutinin mutant SH1-AVPL ( S138A, G186V, T221P, Q226L) from the influenza virus A/Shanghai/1/2013 (H7N9) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Hemagglutinin | | Authors: | Gao, G.F, Xu, Y, Qi, J.X. | | Deposit date: | 2018-09-09 | | Release date: | 2019-11-27 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.383 Å) | | Cite: | Avian-to-Human Receptor-Binding Adaptation of Avian H7N9 Influenza Virus Hemagglutinin.
Cell Rep, 29, 2019
|
|
6ICX
 
 | |
6ID9
 
 | | Crystal structure of H7 hemagglutinin mutant H7-SGTL ( A138S, V186G, P221T) from the influenza virus A/Anhui/1/2013 (H7N9) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Hemagglutinin HA1 chain, Hemagglutinin HA2 chain | | Authors: | Gao, G.F, Xu, Y, Qi, J.X. | | Deposit date: | 2018-09-09 | | Release date: | 2019-11-27 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Avian-to-Human Receptor-Binding Adaptation of Avian H7N9 Influenza Virus Hemagglutinin.
Cell Rep, 29, 2019
|
|
5NJM
 
 | | Lysozyme room-temperature structure determined by serial millisecond crystallography | | Descriptor: | Lysozyme C | | Authors: | Weinert, T, Vera, L, Marsh, M, James, D, Gashi, D, Nogly, P, Jaeger, K, Standfuss, J. | | Deposit date: | 2017-03-29 | | Release date: | 2017-09-27 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Serial millisecond crystallography for routine room-temperature structure determination at synchrotrons.
Nat Commun, 8, 2017
|
|
6DL2
 
 | | BRD4 bromodomain 1 in complex with HYB157 | | Descriptor: | 1,2-ETHANEDIOL, 3-benzyl-2,9-dimethyl-4H,6H-thieno[2,3-e][1,2,4]triazolo[3,4-c][1,4]oxazepine, Bromodomain-containing protein 4 | | Authors: | Meagher, J.L, Stuckey, J.A. | | Deposit date: | 2018-05-31 | | Release date: | 2019-04-17 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Discovery of QCA570 as an Exceptionally Potent and Efficacious Proteolysis Targeting Chimera (PROTAC) Degrader of the Bromodomain and Extra-Terminal (BET) Proteins Capable of Inducing Complete and Durable Tumor Regression.
J. Med. Chem., 61, 2018
|
|
5NM4
 
 | | A2A Adenosine receptor room-temperature structure determined by serial femtosecond crystallography | | Descriptor: | 4-{2-[(7-amino-2-furan-2-yl[1,2,4]triazolo[1,5-a][1,3,5]triazin-5-yl)amino]ethyl}phenol, Adenosine receptor A2a,Soluble cytochrome b562,Adenosine receptor A2a, CHOLESTEROL, ... | | Authors: | Weinert, T, Cheng, R, James, D, Gashi, D, Nogly, P, Jaeger, K, Hennig, M, Standfuss, J. | | Deposit date: | 2017-04-05 | | Release date: | 2017-09-27 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Serial millisecond crystallography for routine room-temperature structure determination at synchrotrons.
Nat Commun, 8, 2017
|
|
5NQT
 
 | | Tubulin Darpin room-temperature structure determined by serial millisecond crystallography | | Descriptor: | DESIGNED ANKYRIN REPEAT PROTEIN (DARPIN) D1, GUANOSINE-5'-DIPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Weinert, T, Olieric, N, James, D, Gashi, D, Nogly, P, Jaeger, K, Steinmetz, M.O, Standfuss, J. | | Deposit date: | 2017-04-21 | | Release date: | 2017-09-27 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Serial millisecond crystallography for routine room-temperature structure determination at synchrotrons.
Nat Commun, 8, 2017
|
|
5NLX
 
 | | A2A Adenosine receptor room-temperature structure determined by serial millisecond crystallography | | Descriptor: | 4-{2-[(7-amino-2-furan-2-yl[1,2,4]triazolo[1,5-a][1,3,5]triazin-5-yl)amino]ethyl}phenol, Adenosine receptor A2a,Soluble cytochrome b562,Adenosine receptor A2a, CHOLESTEROL, ... | | Authors: | Weinert, T, Cheng, R, James, D, Gashi, D, Nogly, P, Jaeger, K, Dore, A.S, Geng, T, Cooke, R, Hennig, M, Standfuss, J. | | Deposit date: | 2017-04-05 | | Release date: | 2017-09-27 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Serial millisecond crystallography for routine room-temperature structure determination at synchrotrons.
Nat Commun, 8, 2017
|
|
5NQU
 
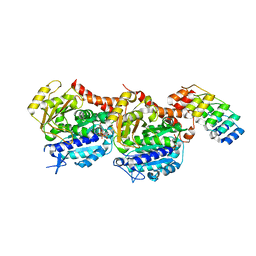 | | Tubulin Darpin cryo structure | | Descriptor: | Designed Ankyrin Repeat Protein (DARPIN) D1, GUANOSINE-5'-DIPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Weinert, T, Olieric, N, James, D, Gashi, D, Nogly, P, Jaeger, K, Steinmetz, M.O, Standfuss, J. | | Deposit date: | 2017-04-21 | | Release date: | 2017-09-27 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Serial millisecond crystallography for routine room-temperature structure determination at synchrotrons.
Nat Commun, 8, 2017
|
|
5O5W
 
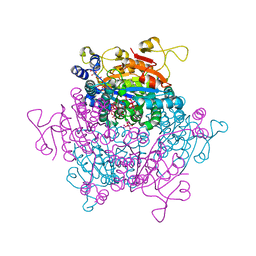 | | Molybdenum storage protein room-temperature structure determined by serial millisecond crystallography | | Descriptor: | (mu3-oxo)-tris(mu2-oxo)-nonakisoxo-trimolybdenum (VI), ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Steffen, B, Weinert, T, Ermler, U, Standfuss, J. | | Deposit date: | 2017-06-02 | | Release date: | 2017-09-27 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Serial millisecond crystallography for routine room-temperature structure determination at synchrotrons.
Nat Commun, 8, 2017
|
|
7BW4
 
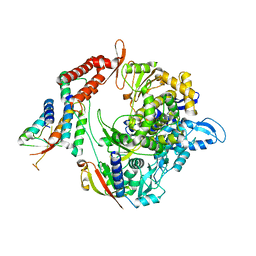 | | Structure of the RNA-dependent RNA polymerase from SARS-CoV-2 | | Descriptor: | Non-structural protein 7, Non-structural protein 8, RNA-directed RNA polymerase, ... | | Authors: | Peng, Q, Peng, R, Shi, Y. | | Deposit date: | 2020-04-13 | | Release date: | 2020-05-27 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Structural and Biochemical Characterization of the nsp12-nsp7-nsp8 Core Polymerase Complex from SARS-CoV-2.
Cell Rep, 31, 2020
|
|
5WQE
 
 | |
5NM2
 
 | | A2A Adenosine receptor cryo structure | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, (2S)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 4-{2-[(7-amino-2-furan-2-yl[1,2,4]triazolo[1,5-a][1,3,5]triazin-5-yl)amino]ethyl}phenol, ... | | Authors: | Weinert, T, Cheng, R, James, D, Gashi, D, Nogly, P, Jaeger, K, Dore, A.S, Geng, T, Cooke, R, Hennig, M, Standfuss, J. | | Deposit date: | 2017-04-05 | | Release date: | 2017-09-27 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.948 Å) | | Cite: | Serial millisecond crystallography for routine room-temperature structure determination at synchrotrons.
Nat Commun, 8, 2017
|
|
5NM5
 
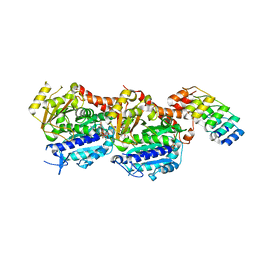 | | Tubulin Darpin room-temperature structure in complex with Colchicine determined by serial millisecond crystallography | | Descriptor: | Designed Ankyrin Repeat Protein (DARPIN) D1, GUANOSINE-5'-DIPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Weinert, T, Olieric, N, James, D, Gashi, D, Nogly, P, Jaeger, K, Steinmetz, M.O, Standfuss, J. | | Deposit date: | 2017-04-05 | | Release date: | 2017-09-27 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Serial millisecond crystallography for routine room-temperature structure determination at synchrotrons.
Nat Commun, 8, 2017
|
|
9ISH
 
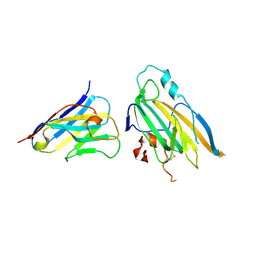 | | Crystal structure of nanobody 32 in complex with HSV-2 gD | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Glycoprotein D, Nanobody 32 | | Authors: | Hu, J, Jin, T.C. | | Deposit date: | 2024-07-17 | | Release date: | 2025-05-28 | | Method: | X-RAY DIFFRACTION (2.88 Å) | | Cite: | A potent protective bispecific nanobody targeting Herpes simplex virus gD reveals vulnerable epitope for neutralizing.
Nat Commun, 16, 2025
|
|
9ISF
 
 | |
1CHZ
 
 | | A NEW NEUROTOXIN FROM BUTHUS MARTENSII KARSCH | | Descriptor: | CHLORIDE ION, PROTEIN (BMK M2) | | Authors: | He, X.L, Deng, J.P, Li, H.M, Wang, D.C. | | Deposit date: | 1999-03-31 | | Release date: | 2000-03-31 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Structure of a new neurotoxin from the scorpion Buthus martensii Karsch at 1.76 A.
Acta Crystallogr.,Sect.D, 56, 2000
|
|
5YAG
 
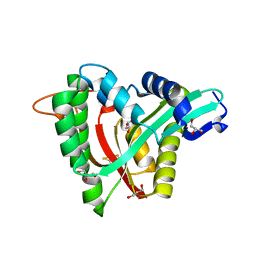 | | Crystal structure of mosquito arylalkylamine N-Acetyltransferase like 5b/spermine N-Acetyltransferase | | Descriptor: | AAEL004827-PA, GLYCEROL | | Authors: | Han, Q, Guan, H, Robinson, H, Li, J. | | Deposit date: | 2017-08-31 | | Release date: | 2018-02-28 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Identification of aaNAT5b as a spermine N-acetyltransferase in the mosquito, Aedes aegypti.
PLoS ONE, 13, 2018
|
|
9BIG
 
 | | Stat6 bound to degrader AK-1690 | | Descriptor: | Signal transducer and activator of transcription 6, [(2-{[(2S)-1-{(2S,4S)-4-[(7-{2-[(3R)-2,6-dioxopiperidin-3-yl]-1-oxo-2,3-dihydro-1H-isoindol-4-yl}hept-6-yn-1-yl)oxy]-2-[(2R)-2-phenylmorpholine-4-carbonyl]pyrrolidin-1-yl}-3,3-dimethyl-1-oxobutan-2-yl]carbamoyl}-1-benzothiophen-5-yl)di(fluoro)methyl]phosphonic acid | | Authors: | Mallik, L, Stuckey, J.A. | | Deposit date: | 2024-04-23 | | Release date: | 2024-10-02 | | Last modified: | 2025-03-26 | | Method: | X-RAY DIFFRACTION (3.304 Å) | | Cite: | Discovery of AK-1690: A Potent and Highly Selective STAT6 PROTAC Degrader.
J.Med.Chem., 68, 2025
|
|
