2PZ9
 
 | | Crystal structure of putative transcriptional regulator SCO4942 from Streptomyces coelicolor | | Descriptor: | Putative regulatory protein, SULFATE ION | | Authors: | Filippova, E.V, Chruszcz, M, Xu, X, Zheng, H, Cymborowski, M, Savchenko, A, Edwards, A, Joachimiak, A, Minor, W, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2007-05-17 | | Release date: | 2007-06-19 | | Last modified: | 2022-04-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | In situ proteolysis for protein crystallization and structure determination.
Nat.Methods, 4, 2007
|
|
1IDY
 
 | |
7CN2
 
 | |
2O8M
 
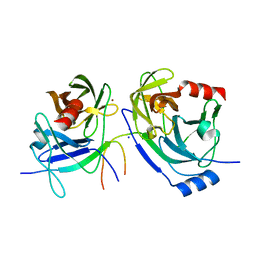 | | Crystal structure of the S139A mutant of Hepatitis C Virus NS3/4A protease | | Descriptor: | Protease, SODIUM ION, ZINC ION | | Authors: | Fischmann, T.O, Prongay, A.J, Madison, V.M, Yao, N. | | Deposit date: | 2006-12-12 | | Release date: | 2007-10-09 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Discovery of the HCV NS3/4A protease inhibitor (1R,5S)-N-[3-amino-1-(cyclobutylmethyl)-2,3-dioxopropyl]-3- [2(S)-[[[(1,1-dimethylethyl)amino]carbonyl]amino]-3,3-dimethyl-1-oxobutyl]- 6,6-dimethyl-3-azabicyclo[3.1.0]hexan-2(S)-carboxamide (Sch 503034) II. Key steps in structure-based optimization
J.Med.Chem., 50, 2007
|
|
5GNV
 
 | | Structure of PSD-95/MAP1A complex reveals unique target recognition mode of MAGUK GK domain | | Descriptor: | Disks large homolog 4, Microtubule-associated protein 1A, SULFATE ION | | Authors: | Shang, Y, Xia, Y, Zhu, R, Zhu, J. | | Deposit date: | 2016-07-25 | | Release date: | 2017-08-02 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.596 Å) | | Cite: | Structure of the PSD-95/MAP1A complex reveals a unique target recognition mode of the MAGUK GK domain
Biochem. J., 474, 2017
|
|
8JHF
 
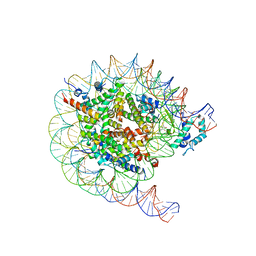 | | Native SUV420H1 bound to 167-bp nucleosome | | Descriptor: | DNA (160-MER), Histone H2A.Z, Histone H2B type 1-K, ... | | Authors: | Lin, F, Li, W. | | Deposit date: | 2023-05-23 | | Release date: | 2023-11-15 | | Last modified: | 2023-12-20 | | Method: | ELECTRON MICROSCOPY (3.68 Å) | | Cite: | Structural basis of nucleosomal H4K20 recognition and methylation by SUV420H1 methyltransferase.
Cell Discov, 9, 2023
|
|
8JHG
 
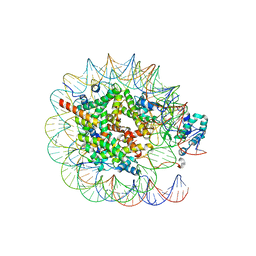 | | Native SUV420H1 bound to 167-bp nucleosome | | Descriptor: | DNA (160-MER), Histone H2A type 1-B/E, Histone H2B type 1-K, ... | | Authors: | Lin, F, Li, W. | | Deposit date: | 2023-05-23 | | Release date: | 2023-11-22 | | Last modified: | 2023-12-20 | | Method: | ELECTRON MICROSCOPY (3.58 Å) | | Cite: | Structural basis of nucleosomal H4K20 recognition and methylation by SUV420H1 methyltransferase.
Cell Discov, 9, 2023
|
|
7XS8
 
 | |
7XSC
 
 | |
7XSA
 
 | |
7XSB
 
 | |
4FIH
 
 | |
4FIJ
 
 | | Catalytic domain of human PAK4 | | Descriptor: | Serine/threonine-protein kinase PAK 4 | | Authors: | Ha, B.H, Boggon, T.J. | | Deposit date: | 2012-06-08 | | Release date: | 2012-09-12 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Type II p21-activated kinases (PAKs) are regulated by an autoinhibitory pseudosubstrate.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
4FIE
 
 | | Full-length human PAK4 | | Descriptor: | PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, Serine/threonine-protein kinase PAK 4 | | Authors: | Ha, B.H, Boggon, T.J. | | Deposit date: | 2012-06-08 | | Release date: | 2012-09-12 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3.11 Å) | | Cite: | Type II p21-activated kinases (PAKs) are regulated by an autoinhibitory pseudosubstrate.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
4FIG
 
 | | Catalytic domain of human PAK4 | | Descriptor: | MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, Serine/threonine-protein kinase PAK 4 | | Authors: | Ha, B.H, Boggon, T.J. | | Deposit date: | 2012-06-08 | | Release date: | 2012-09-12 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3.01 Å) | | Cite: | Type II p21-activated kinases (PAKs) are regulated by an autoinhibitory pseudosubstrate.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
4FIF
 
 | | Catalytic domain of human PAK4 with RPKPLVDP peptide | | Descriptor: | PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, Serine/threonine-protein kinase PAK 4 | | Authors: | Ha, B.H, Boggon, T.J. | | Deposit date: | 2012-06-08 | | Release date: | 2012-09-12 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Type II p21-activated kinases (PAKs) are regulated by an autoinhibitory pseudosubstrate.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
2QMK
 
 | | Human pancreatic alpha-amylase complexed with nitrite | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, NITRITE ION, ... | | Authors: | Williams, L.K, Maurus, R, Brayer, G.D. | | Deposit date: | 2007-07-16 | | Release date: | 2008-03-25 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Alternative catalytic anions differentially modulate human alpha-amylase activity and specificity
Biochemistry, 47, 2008
|
|
4FII
 
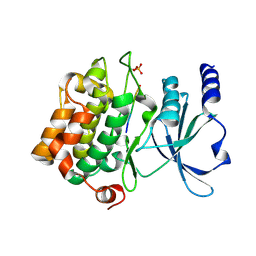 | | Catalytic domain of human PAK4 with RPKPLVDP peptide | | Descriptor: | Serine/threonine-protein kinase PAK 4 | | Authors: | Ha, B.H, Boggon, T.J. | | Deposit date: | 2012-06-08 | | Release date: | 2012-09-12 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Type II p21-activated kinases (PAKs) are regulated by an autoinhibitory pseudosubstrate.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
1IDZ
 
 | |
2QV4
 
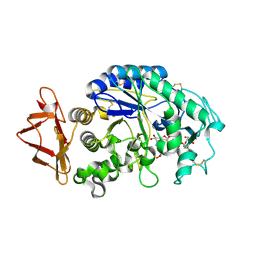 | | Human pancreatic alpha-amylase complexed with nitrite and acarbose | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 4,6-dideoxy-4-{[(1S,4R,5R,6S)-4-{[alpha-D-glucopyranosyl-(1->4)-alpha-D-glucopyranosyl-(1->4)-alpha-D-glucopyranosyl]oxy}-5,6-dihydroxy-3-(hydroxymethyl)cyclohex-2-en-1-yl]amino}-alpha-D-glucopyranose, CALCIUM ION, ... | | Authors: | Williams, L.K, Maurus, R, Brayer, G.D. | | Deposit date: | 2007-08-07 | | Release date: | 2008-03-25 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Alternative catalytic anions differentially modulate human alpha-amylase activity and specificity
Biochemistry, 47, 2008
|
|
2ERF
 
 | |
2BBH
 
 | | X-ray structure of T.maritima CorA soluble domain | | Descriptor: | DECYL-BETA-D-MALTOPYRANOSIDE, MAGNESIUM ION, SODIUM ION, ... | | Authors: | Lunin, V.V, Dobrovetsky, E, Khutoreskaya, G, Bochkarev, A, Maguire, M.E, Edwards, A.M, Koth, C.M, Structural Genomics Consortium (SGC) | | Deposit date: | 2005-10-17 | | Release date: | 2005-12-13 | | Last modified: | 2017-10-18 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal structure of the CorA Mg2+ transporter.
Nature, 440, 2006
|
|
3HAD
 
 | |
6IM6
 
 | | Crystal structure of PDE4D complexed with a novel inhibitor | | Descriptor: | 1,2-ETHANEDIOL, 7-ethoxy-6-methoxy-3,4-dihydroisoquinoline-2(1H)-carbaldehyde, MAGNESIUM ION, ... | | Authors: | Zhang, X.L, Su, H.X, Xu, Y.C. | | Deposit date: | 2018-10-22 | | Release date: | 2019-10-23 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.702 Å) | | Cite: | Structure-Aided Identification and Optimization of Tetrahydro-isoquinolines as Novel PDE4 Inhibitors Leading to Discovery of an Effective Antipsoriasis Agent.
J.Med.Chem., 62, 2019
|
|
6IMT
 
 | | Crystal structure of PDE4D complexed with a novel inhibitor | | Descriptor: | (1S)-1-[2-(6-fluoro-1H-indol-3-yl)ethyl]-6,7-dimethoxy-3,4-dihydroisoquinoline-2(1H)-carbaldehyde, 1,2-ETHANEDIOL, MAGNESIUM ION, ... | | Authors: | Zhang, X.L, Su, H.X, Xu, Y.C. | | Deposit date: | 2018-10-23 | | Release date: | 2019-10-23 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.483 Å) | | Cite: | Structure-Aided Identification and Optimization of Tetrahydro-isoquinolines as Novel PDE4 Inhibitors Leading to Discovery of an Effective Antipsoriasis Agent.
J.Med.Chem., 62, 2019
|
|
