4TW0
 
 | | Crystal Structure of SCARB2 in Acidic Condition (pH4.8) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Dang, M.H, Wang, X.X, Rao, Z.H. | | Deposit date: | 2014-06-29 | | Release date: | 2015-07-08 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (3.648 Å) | | Cite: | Molecular mechanism of SCARB2-mediated attachment and uncoating of EV71
Protein Cell, 5, 2014
|
|
3URF
 
 | | Human RANKL/OPG complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Tumor necrosis factor ligand superfamily member 11, soluble form, ... | | Authors: | Wang, X.Q, Luan, X.D, Lu, Q.Y. | | Deposit date: | 2011-11-22 | | Release date: | 2012-07-11 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.701 Å) | | Cite: | Crystal Structure of Human RANKL Complexed with Its Decoy Receptor Osteoprotegerin
J.Immunol., 189, 2012
|
|
4NJL
 
 | | Crystal structure of middle east respiratory syndrome coronavirus S2 protein fusion core | | Descriptor: | S protein, TRIETHYLENE GLYCOL | | Authors: | Zhu, Y, Lu, L, Qin, L, Ye, S, Jiang, S, Zhang, R. | | Deposit date: | 2013-11-10 | | Release date: | 2014-02-19 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure-based discovery of Middle East respiratory syndrome coronavirus fusion inhibitor.
Nat Commun, 5, 2014
|
|
3ZID
 
 | |
3S4S
 
 | | Crystal structure of CD4 mutant bound to HLA-DR1 | | Descriptor: | GLYCEROL, HLA class II histocompatibility antigen, DR alpha chain, ... | | Authors: | Li, Y. | | Deposit date: | 2011-05-20 | | Release date: | 2011-09-21 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Affinity maturation of human CD4 by yeast surface display and crystal structure of a CD4-HLA-DR1 complex.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
7QI8
 
 | | CRYSTAL STRUCTURE OF LYSYL-TRNA SYNTHETASE FROM Mycobacterium tuberculosis COMPLEXED WITH L-LYSINE AND INHIBITOR | | Descriptor: | 2-azanyl-6-[(1~{S},7~{S})-2,2-bis(fluoranyl)-7-oxidanyl-cycloheptyl]-4-methoxy-7~{H}-pyrrolo[3,4-d]pyrimidin-5-one, LYSINE, Lysine--tRNA ligase 1 | | Authors: | Dawson, A, Robinson, D.A, Tamjar, J, Wyatt, P, Green, S. | | Deposit date: | 2021-12-14 | | Release date: | 2022-10-19 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Lysyl-tRNA synthetase, a target for urgently needed M. tuberculosis drugs.
Nat Commun, 13, 2022
|
|
7QHN
 
 | | CRYSTAL STRUCTURE OF LYSYL-TRNA SYNTHETASE FROM Mycobacterium tuberculosis COMPLEXED WITH L-LYSINE and an inhibitor | | Descriptor: | 6-azanyl-2-cyclohexyl-4-fluoranyl-1~{H}-pyrrolo[3,4-c]pyridin-3-one, LYSINE, Lysine--tRNA ligase 1 | | Authors: | Dawson, A, Robinson, D.A, Tamjar, J, Wyatt, P, Green, S. | | Deposit date: | 2021-12-13 | | Release date: | 2022-10-19 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.58 Å) | | Cite: | Lysyl-tRNA synthetase, a target for urgently needed M. tuberculosis drugs.
Nat Commun, 13, 2022
|
|
7QH8
 
 | | CRYSTAL STRUCTURE OF LYSYL-TRNA SYNTHETASE FROM Mycobacterium tuberculosis COMPLEXED WITH L-LYSINE | | Descriptor: | GLYCEROL, LYSINE, Lysine--tRNA ligase 1 | | Authors: | Dawson, A, Robinson, D.A, Tamjar, J, Wyatt, P, Green, S. | | Deposit date: | 2021-12-10 | | Release date: | 2022-10-19 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Lysyl-tRNA synthetase, a target for urgently needed M. tuberculosis drugs.
Nat Commun, 13, 2022
|
|
4A2B
 
 | |
4A2A
 
 | | Thermotoga maritima FtsA:FtsZ(336-351) | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, CELL DIVISION PROTEIN FTSA, PUTATIVE, ... | | Authors: | Szwedziak, P, Lowe, J. | | Deposit date: | 2011-09-23 | | Release date: | 2012-04-25 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Ftsa Forms Actin-Like Protofilaments
Embo J., 31, 2012
|
|
6WO1
 
 | |
4QR4
 
 | | Brd4 Bromodomain 1 complex with its novel inhibitors | | Descriptor: | 2-chloro-N-cyclopentyl-5-(2-oxo-2,3-dihydro-1,3-thiazol-4-yl)benzenesulfonamide, Bromodomain-containing protein 4 | | Authors: | Xiong, B, Cao, D.Y, Chen, T.T, Xu, Y.C. | | Deposit date: | 2014-06-30 | | Release date: | 2015-07-01 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.28 Å) | | Cite: | Fragment-based drug discovery of 2-thiazolidinones as BRD4 inhibitors: 2. Structure-based optimization
J.Med.Chem., 58, 2015
|
|
5NEM
 
 | |
6U9H
 
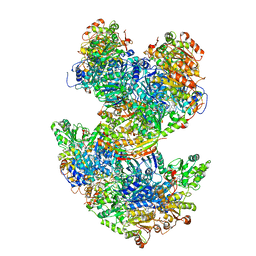 | | Arabidopsis thaliana acetohydroxyacid synthase complex | | Descriptor: | Acetolactate synthase small subunit 2, chloroplastic, Acetolactate synthase, ... | | Authors: | Guddat, L.W, Low, Y.S, Rao, Z. | | Deposit date: | 2019-09-08 | | Release date: | 2020-07-15 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structures of fungal and plant acetohydroxyacid synthases.
Nature, 586, 2020
|
|
5NED
 
 | |
5OC9
 
 | | Crystal Structure of human TMEM16K / Anoctamin 10 | | Descriptor: | (2R)-2,3-dihydroxypropyl (7Z)-hexadec-7-enoate, Anoctamin-10, CALCIUM ION | | Authors: | Bushell, S.R, Pike, A.C.W, Chu, A, Tessitore, A, Rotty, B, Mukhopadhyay, S, Kupinska, K, Shrestha, L, Borkowska, O, Chalk, R, Burgess-Brown, N.A, Love, J, Edwards, A.M, Arrowsmith, C.H, Bountra, C, Carpenter, E.P, Structural Genomics Consortium (SGC) | | Deposit date: | 2017-06-29 | | Release date: | 2018-07-25 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | The structural basis of lipid scrambling and inactivation in the endoplasmic reticulum scramblase TMEM16K.
Nat Commun, 10, 2019
|
|
5NEJ
 
 | |
5NET
 
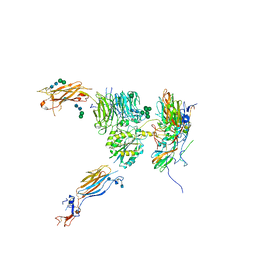 | |
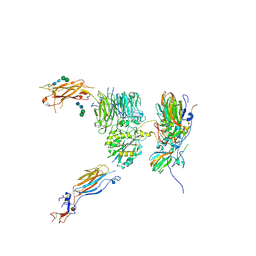
5NER
 
 | |
3S5L
 
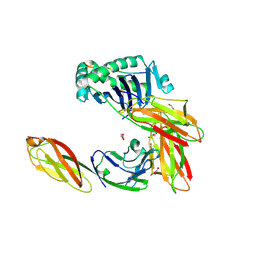 | | Crystal structure of CD4 mutant bound to HLA-DR1 | | Descriptor: | GLYCEROL, HA peptide, HLA class II histocompatibility antigen DR beta chain, ... | | Authors: | Li, Y. | | Deposit date: | 2011-05-23 | | Release date: | 2011-09-21 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Affinity maturation of human CD4 by yeast surface display and crystal structure of a CD4-HLA-DR1 complex.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
7C2Q
 
 | | The crystal structure of COVID-19 main protease in the apo state | | Descriptor: | 3C-like proteinase | | Authors: | Zhou, X.L, Zhong, F.L, Lin, C, Hu, X.H, Zhou, H, Wang, Q.S, Li, j, Zhang, J. | | Deposit date: | 2020-05-08 | | Release date: | 2020-09-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Structure of SARS-CoV-2 main protease in the apo state.
Sci China Life Sci, 64, 2021
|
|
6U9D
 
 | |
8HWT
 
 | | SARS-CoV-2 Omicron BA.2 RBD complexed with BD-604 and S304 Fab | | Descriptor: | BD-604 heavy chain, BD-604 light chain, S304 heavy chain, ... | | Authors: | He, Q.W, Xie, Y. | | Deposit date: | 2023-01-02 | | Release date: | 2023-12-06 | | Last modified: | 2025-07-16 | | Method: | ELECTRON MICROSCOPY (2.91 Å) | | Cite: | An updated atlas of antibody evasion by SARS-CoV-2 Omicron sub-variants including BQ.1.1 and XBB.
Cell Rep Med, 4, 2023
|
|
8HWS
 
 | | The complex structure of Omicron BA.4 RBD with BD604, S309, and S304 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, BD-604 Fab Heavy chain, BD-604 Fab Light chain, ... | | Authors: | He, Q.W, Xu, Z.P, Xie, Y.F. | | Deposit date: | 2023-01-02 | | Release date: | 2023-12-06 | | Last modified: | 2025-07-16 | | Method: | ELECTRON MICROSCOPY (2.36 Å) | | Cite: | An updated atlas of antibody evasion by SARS-CoV-2 Omicron sub-variants including BQ.1.1 and XBB.
Cell Rep Med, 4, 2023
|
|
4B45
 
 | |
