6N44
 
 | |
3UTS
 
 | | 1E6-A*0201-ALWGPDPAAA Complex, Monoclinic | | Descriptor: | 1E6 TCR Alpha Chain, 1E6 TCR Beta Chain, Beta-2-microglobulin, ... | | Authors: | Rizkallah, P.J, Cole, D.K, Sewell, A.K, Bulek, A.M. | | Deposit date: | 2011-11-26 | | Release date: | 2012-01-25 | | Last modified: | 2012-03-07 | | Method: | X-RAY DIFFRACTION (2.712 Å) | | Cite: | Structural basis for the killing of human beta cells by CD8(+) T cells in type 1 diabetes.
Nat.Immunol., 13, 2012
|
|
2PEE
 
 | | Crystal Structure of a Thermophilic Serpin, Tengpin, in the Native State | | Descriptor: | GLYCEROL, SULFATE ION, Serine protease inhibitor | | Authors: | Zhang, Q.W, Buckle, A.M, Whisstock, J.C. | | Deposit date: | 2007-04-02 | | Release date: | 2007-06-19 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | The N terminus of the serpin, tengpin, functions to trap the metastable native state.
Embo Rep., 8, 2007
|
|
2NW0
 
 | | Crystal structure of a lysin | | Descriptor: | ACETATE ION, PlyB | | Authors: | Porter, C.J, Buckle, A.M, Whisstock, J.C. | | Deposit date: | 2006-11-14 | | Release date: | 2006-12-05 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The 1.6 A Crystal Structure of the Catalytic Domain of PlyB, a Bacteriophage Lysin Active Against Bacillus anthracis.
J.Mol.Biol., 366, 2007
|
|
3HUJ
 
 | |
3UTP
 
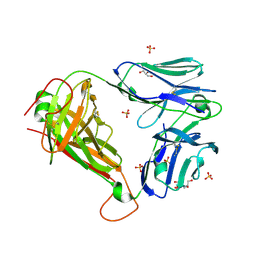 | | 1E6 TCR specific for HLA-A*0201-ALWGPDPAAA | | Descriptor: | 1E6 TCR alpha chain, 1E6 TCR beta chain, 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ... | | Authors: | Rizkallah, P.J, Cole, D.K, Sewell, A.K, Bulek, A.M. | | Deposit date: | 2011-11-26 | | Release date: | 2012-01-25 | | Last modified: | 2012-03-07 | | Method: | X-RAY DIFFRACTION (2.574 Å) | | Cite: | Structural basis for the killing of human beta cells by CD8(+) T cells in type 1 diabetes.
Nat.Immunol., 13, 2012
|
|
2PEF
 
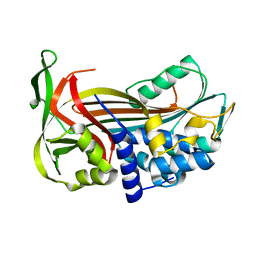 | | Crystal Structure of a Thermophilic Serpin, Tengpin, in the Latent State | | Descriptor: | Serine protease inhibitor | | Authors: | Zhang, Q.W, Buckle, A.M, Whisstock, J.C. | | Deposit date: | 2007-04-03 | | Release date: | 2007-06-19 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The N terminus of the serpin, tengpin, functions to trap the metastable native state.
Embo Rep., 8, 2007
|
|
3UTQ
 
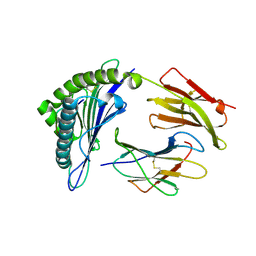 | | Human HLA-A*0201-ALWGPDPAAA | | Descriptor: | Beta-2-microglobulin, HLA class I histocompatibility antigen, A-2 alpha chain, ... | | Authors: | Rizkallah, P.J, Cole, D.K, Sewell, A.K, Bulek, A.M. | | Deposit date: | 2011-11-26 | | Release date: | 2012-01-25 | | Last modified: | 2012-03-07 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Structural basis for the killing of human beta cells by CD8(+) T cells in type 1 diabetes.
Nat.Immunol., 13, 2012
|
|
6VO1
 
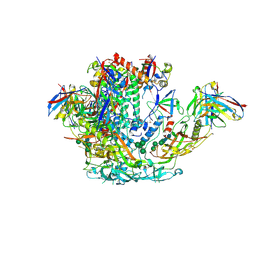 | | BG505 SOSIP.v5.2 in complex with rhesus macaque Fab RM20J | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Envelope glycoprotein gp120, ... | | Authors: | Cottrell, C.A, Ward, A.B. | | Deposit date: | 2020-01-29 | | Release date: | 2020-07-01 | | Last modified: | 2020-09-16 | | Method: | ELECTRON MICROSCOPY (3.88 Å) | | Cite: | Mapping the immunogenic landscape of near-native HIV-1 envelope trimers in non-human primates.
Plos Pathog., 16, 2020
|
|
6VLR
 
 | | BG505 SOSIP.v5.2 in complex with rhesus macaque Fab RM20E1 and PGT122 Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, BG505 SOSIPv5.2 gp41, ... | | Authors: | Cottrell, C.A, Ward, A.B. | | Deposit date: | 2020-01-24 | | Release date: | 2020-06-24 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (4.42 Å) | | Cite: | Mapping the immunogenic landscape of near-native HIV-1 envelope trimers in non-human primates.
Plos Pathog., 16, 2020
|
|
6VN0
 
 | | BG505 SOSIP.v4.1 in complex with rhesus macaque Fab RM20F | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Envelope glycoprotein gp160, ... | | Authors: | Cottrell, C.A, Shin, M, Ward, A.B. | | Deposit date: | 2020-01-29 | | Release date: | 2020-06-24 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (4.25 Å) | | Cite: | Mapping the immunogenic landscape of near-native HIV-1 envelope trimers in non-human primates.
Plos Pathog., 16, 2020
|
|
7GSS
 
 | | Human glutathione S-transferase P1-1, complex with glutathione | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, GLUTATHIONE, GLUTATHIONE S-TRANSFERASE P1-1 | | Authors: | Oakley, A, Parker, M. | | Deposit date: | 1997-08-13 | | Release date: | 1998-09-16 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The structures of human glutathione transferase P1-1 in complex with glutathione and various inhibitors at high resolution.
J.Mol.Biol., 274, 1997
|
|
20GS
 
 | | GLUTATHIONE S-TRANSFERASE P1-1 COMPLEXED WITH CIBACRON BLUE | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CIBACRON BLUE, GLUTATHIONE S-TRANSFERASE | | Authors: | Oakley, A.J, Lo Bello, M, Nuccetelli, M, Mazzetti, A.P, Parker, M.W. | | Deposit date: | 1997-12-16 | | Release date: | 1998-12-30 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | The ligandin (non-substrate) binding site of human Pi class glutathione transferase is located in the electrophile binding site (H-site).
J.Mol.Biol., 291, 1999
|
|
22GS
 
 | | HUMAN GLUTATHIONE S-TRANSFERASE P1-1 Y49F MUTANT | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, GLUTATHIONE S-TRANSFERASE P1-1 | | Authors: | Oakley, A.J. | | Deposit date: | 1998-03-10 | | Release date: | 1999-03-23 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Thermodynamic description of the effect of the mutation Y49F on human glutathione transferase P1-1 in binding with glutathione and the inhibitor S-hexylglutathione.
J.Biol.Chem., 278, 2003
|
|
2BK2
 
 | | The prepore structure of pneumolysin, obtained by fitting the alpha carbon trace of perfringolysin O into a cryo-EM map | | Descriptor: | PERFRINGOLYSIN O | | Authors: | Tilley, S.J, Orlova, E.V, Gilbert, R.J.C, Andrew, P.W, Saibil, H.R. | | Deposit date: | 2005-02-10 | | Release date: | 2005-05-04 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (28 Å) | | Cite: | Structural Basis of Pore Formation by the Bacterial Toxin Pneumolysin
Cell(Cambridge,Mass.), 121, 2005
|
|
8CON
 
 | | Crystal structure of alcohol dehydrogenase from Arabidopsis thaliana in complex with NADH | | Descriptor: | 1,2-ETHANEDIOL, 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, Alcohol dehydrogenase class-P, ... | | Authors: | Fermani, S, Fanti, S, Carloni, G, Falini, G, Meloni, M, Zaffagnini, M. | | Deposit date: | 2023-02-28 | | Release date: | 2024-02-21 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural and biochemical characterization of Arabidopsis alcohol dehydrogenases reveals distinct functional properties but similar redox sensitivity.
Plant J., 118, 2024
|
|
7R75
 
 | | Structure of human SHP2 in complex with compound 16 | | Descriptor: | 6-(4-amino-4-methylpiperidin-1-yl)-3-(3-chlorophenyl)-1,5-dihydro-4H-pyrazolo[3,4-d]pyrimidin-4-one, Tyrosine-protein phosphatase non-receptor type 11 | | Authors: | Leonard, P.G, Cross, J. | | Deposit date: | 2021-06-24 | | Release date: | 2021-10-27 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.83 Å) | | Cite: | Discovery of 6-[(3 S ,4 S )-4-Amino-3-methyl-2-oxa-8-azaspiro[4.5]decan-8-yl]-3-(2,3-dichlorophenyl)-2-methyl-3,4-dihydropyrimidin-4-one (IACS-15414), a Potent and Orally Bioavailable SHP2 Inhibitor.
J.Med.Chem., 64, 2021
|
|
7R7D
 
 | | Structure of human SHP2 in complex with compound 22 | | Descriptor: | 4-[6-(4-amino-4-methylpiperidin-1-yl)-1H-pyrazolo[3,4-b]pyrazin-3-yl]-3-chloro-N-methylpyridin-2-amine, TETRAETHYLENE GLYCOL, Tyrosine-protein phosphatase non-receptor type 11 | | Authors: | Leonard, P.G, Cross, J. | | Deposit date: | 2021-06-24 | | Release date: | 2021-10-27 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Discovery of 6-[(3 S ,4 S )-4-Amino-3-methyl-2-oxa-8-azaspiro[4.5]decan-8-yl]-3-(2,3-dichlorophenyl)-2-methyl-3,4-dihydropyrimidin-4-one (IACS-15414), a Potent and Orally Bioavailable SHP2 Inhibitor.
J.Med.Chem., 64, 2021
|
|
7R7L
 
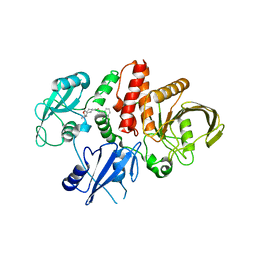 | | Structure of human SHP2 in complex with compound 30 | | Descriptor: | 6-[(3S,4S)-4-amino-3-methyl-2-oxa-8-azaspiro[4.5]decan-8-yl]-3-(2,3-dichlorophenyl)-2-methylpyrimidin-4(3H)-one, Tyrosine-protein phosphatase non-receptor type 11 | | Authors: | Leonard, P.G, Cross, J. | | Deposit date: | 2021-06-24 | | Release date: | 2021-10-27 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Discovery of 6-[(3 S ,4 S )-4-Amino-3-methyl-2-oxa-8-azaspiro[4.5]decan-8-yl]-3-(2,3-dichlorophenyl)-2-methyl-3,4-dihydropyrimidin-4-one (IACS-15414), a Potent and Orally Bioavailable SHP2 Inhibitor.
J.Med.Chem., 64, 2021
|
|
7R7I
 
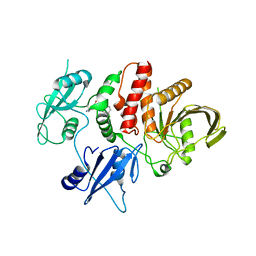 | | Structure of human SHP2 in complex with compound 27 | | Descriptor: | Tyrosine-protein phosphatase non-receptor type 11, [3-(4-amino-4-methylpiperidin-1-yl)-6-(2,3-dichlorophenyl)-5-methylpyrazin-2-yl]methanol | | Authors: | Leonard, P.G, Cross, J. | | Deposit date: | 2021-06-24 | | Release date: | 2021-10-27 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Discovery of 6-[(3 S ,4 S )-4-Amino-3-methyl-2-oxa-8-azaspiro[4.5]decan-8-yl]-3-(2,3-dichlorophenyl)-2-methyl-3,4-dihydropyrimidin-4-one (IACS-15414), a Potent and Orally Bioavailable SHP2 Inhibitor.
J.Med.Chem., 64, 2021
|
|
6NF7
 
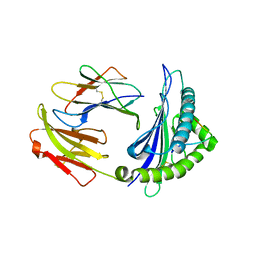 | | Crystal Structure of RT1.Aa-Bu31-10 | | Descriptor: | Beta-2-microglobulin, Bu31-10 peptide, RT1A.a | | Authors: | Gras, S. | | Deposit date: | 2018-12-19 | | Release date: | 2019-12-25 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Cross-Reactive Donor-Specific CD8+Tregs Efficiently Prevent Transplant Rejection.
Cell Rep, 29, 2019
|
|
7SNE
 
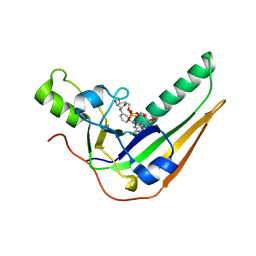 | | Pertussis toxin S1 subunit bound to BaAD | | Descriptor: | Pertussis toxin subunit 1, [(2R,3S,4R,5R)-5-(6-amino-9H-purin-9-yl)-3,4-dihydroxyoxolan-2-yl]methyl [(2R,3S,4R,5S)-5-(3-carbamoylanilino)-3,4-dihydroxyoxolan-2-yl]methyl dihydrogen diphosphate (non-preferred name) | | Authors: | Littler, D.R, Beddoe, T. | | Deposit date: | 2021-10-28 | | Release date: | 2022-04-13 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.00011 Å) | | Cite: | Crystal structures of pertussis toxin with NAD + and analogs provide structural insights into the mechanism of its cytosolic ADP-ribosylation activity.
J.Biol.Chem., 298, 2022
|
|
7T5M
 
 | | Structure of HLA-A*02:01-FLPTPEELGLLGPPRPQVLA complex | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Beta-2-microglobulin, Interleukin-27 receptor subunit alpha, ... | | Authors: | Gras, S. | | Deposit date: | 2021-12-12 | | Release date: | 2022-12-14 | | Last modified: | 2024-01-03 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Cutting Edge: Unconventional CD8 + T Cell Recognition of a Naturally Occurring HLA-A*02:01-Restricted 20mer Epitope.
J Immunol., 208, 2022
|
|
7T0L
 
 | |
6MT3
 
 | |
