7WMJ
 
 | | A novel chemical derivative(71) of THRB agonist | | Descriptor: | 2-[[7-[2,6-dimethyl-4-(phenylcarbonyl)phenoxy]-1-methoxy-4-oxidanyl-isoquinolin-3-yl]carbonylamino]ethanoic acid, Isoform Beta-2 of Thyroid hormone receptor beta, Nuclear receptor coactivator 2 | | Authors: | Yao, B.Q, Li, Y. | | Deposit date: | 2022-01-14 | | Release date: | 2022-05-18 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.81 Å) | | Cite: | Discovery of a Highly Selective and H435R-Sensitive Thyroid Hormone Receptor beta Agonist.
J.Med.Chem., 65, 2022
|
|
7WMH
 
 | | A novel chemical derivative(56) of THRB agonist | | Descriptor: | 2-[[1-methoxy-4-oxidanyl-7-(4-phenylphenoxy)isoquinolin-3-yl]carbonylamino]ethanoic acid, Isoform Beta-2 of Thyroid hormone receptor beta, Nuclear receptor coactivator 2 | | Authors: | Yao, B.Q, Li, Y. | | Deposit date: | 2022-01-14 | | Release date: | 2022-05-18 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Discovery of a Highly Selective and H435R-Sensitive Thyroid Hormone Receptor beta Agonist.
J.Med.Chem., 65, 2022
|
|
7WMG
 
 | | A novel chemical derivative(52) of THRB agonist | | Descriptor: | 2-[[1-methoxy-4-oxidanyl-7-(4-phenoxyphenoxy)isoquinolin-3-yl]carbonylamino]ethanoic acid, Isoform Beta-2 of Thyroid hormone receptor beta, Nuclear receptor coactivator 2 | | Authors: | Yao, B.Q, Li, Y. | | Deposit date: | 2022-01-14 | | Release date: | 2022-05-18 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.93 Å) | | Cite: | Discovery of a Highly Selective and H435R-Sensitive Thyroid Hormone Receptor beta Agonist.
J.Med.Chem., 65, 2022
|
|
7WMN
 
 | | A novel chemical derivative(89) of THRB agonist | | Descriptor: | 2-[[7-[2,6-dimethyl-4-(3-methylphenyl)carbonyl-phenoxy]-1-methoxy-4-oxidanyl-isoquinolin-3-yl]carbonylamino]ethanoic acid, Isoform Beta-2 of Thyroid hormone receptor beta, Nuclear receptor coactivator 2 | | Authors: | Yao, B.Q, Li, Y. | | Deposit date: | 2022-01-15 | | Release date: | 2022-05-18 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.57 Å) | | Cite: | Discovery of a Highly Selective and H435R-Sensitive Thyroid Hormone Receptor beta Agonist.
J.Med.Chem., 65, 2022
|
|
7WLX
 
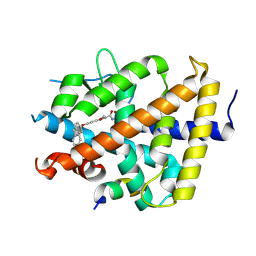 | | A novel chemical derivative(53) of THRB agonist | | Descriptor: | 2-[[1-methoxy-4-oxidanyl-7-[4-(phenylmethyl)phenoxy]isoquinolin-3-yl]carbonylamino]ethanoic acid, Isoform Beta-2 of Thyroid hormone receptor beta, Nuclear receptor coactivator 2 | | Authors: | Yao, B.Q, Li, Y. | | Deposit date: | 2022-01-14 | | Release date: | 2022-05-18 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | Discovery of a Highly Selective and H435R-Sensitive Thyroid Hormone Receptor beta Agonist.
J.Med.Chem., 65, 2022
|
|
8HZQ
 
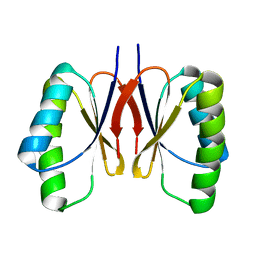 | | Bacillus subtilis SepF protein assembly (wild type) | | Descriptor: | Cell division protein SepF | | Authors: | Liu, W. | | Deposit date: | 2023-01-09 | | Release date: | 2024-02-14 | | Last modified: | 2024-05-15 | | Method: | SOLID-STATE NMR | | Cite: | Molecular basis for curvature formation in SepF polymerization.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
8HZT
 
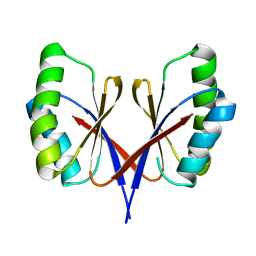 | |
5TUG
 
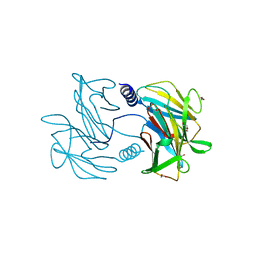 | |
5TUH
 
 | |
7XPN
 
 | |
7YP3
 
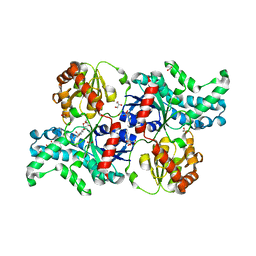 | | Crystal structure of elaiophylin glycosyltransferase in complex with elaiophylin | | Descriptor: | ACETATE ION, Elaiophylin, GLYCEROL, ... | | Authors: | Xu, T, Liu, Q, Gan, Q, Liu, J. | | Deposit date: | 2022-08-02 | | Release date: | 2022-11-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Substrate-induced dimerization of elaiophylin glycosyltransferase reveals a novel self-activating form of glycosyltransferase for symmetric glycosylation.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
7YP6
 
 | | Crystal structure of elaiophylin glycosyltransferase in complex with UDP | | Descriptor: | Glycosyltransferase, R-1,2-PROPANEDIOL, URIDINE-5'-DIPHOSPHATE | | Authors: | Xu, T, Liu, Q, Gan, Q, Liu, J. | | Deposit date: | 2022-08-02 | | Release date: | 2022-11-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Substrate-induced dimerization of elaiophylin glycosyltransferase reveals a novel self-activating form of glycosyltransferase for symmetric glycosylation.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
7YP5
 
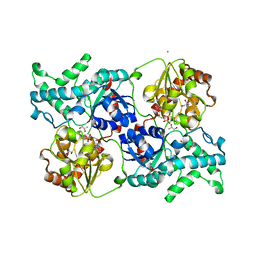 | | Crystal structure of elaiophylin glycosyltransferase in complex with TDP | | Descriptor: | CHLORIDE ION, Glycosyltransferase, R-1,2-PROPANEDIOL, ... | | Authors: | Xu, T, Liu, Q, Gan, Q, Liu, J. | | Deposit date: | 2022-08-02 | | Release date: | 2022-11-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | Substrate-induced dimerization of elaiophylin glycosyltransferase reveals a novel self-activating form of glycosyltransferase for symmetric glycosylation.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
7YP4
 
 | | Crystal structure of elaiophylin glycosyltransferase in apo-form | | Descriptor: | Glycosyltransferase, R-1,2-PROPANEDIOL | | Authors: | Xu, T, Liu, Q, Gan, Q, Liu, J. | | Deposit date: | 2022-08-02 | | Release date: | 2022-11-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Substrate-induced dimerization of elaiophylin glycosyltransferase reveals a novel self-activating form of glycosyltransferase for symmetric glycosylation.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
7DHJ
 
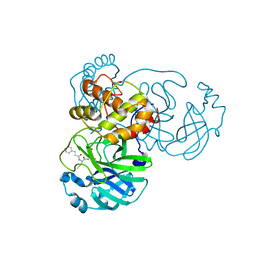 | | The co-crystal structure of SARS-CoV-2 main protease with the peptidomimetic inhibitor (S)-2-cinnamamido-N-((S)-1-oxo-3-((S)-2-oxopyrrolidin-3-yl)propan-2-yl)pent-4-ynamide | | Descriptor: | (2~{S})-~{N}-[(2~{S})-1-oxidanylidene-3-[(3~{S})-2-oxidanylidenepyrrolidin-3-yl]propan-2-yl]-2-[[(~{E})-3-phenylprop-2-enoyl]amino]pent-4-ynamide, 3C-like proteinase | | Authors: | Shang, L.Q, Wang, H, Deng, W.L, Xing, S, Wang, Y.X. | | Deposit date: | 2020-11-15 | | Release date: | 2021-11-24 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.962 Å) | | Cite: | The structure-based design of peptidomimetic inhibitors against SARS-CoV-2 3C like protease as Potent anti-viral drug candidate.
Eur.J.Med.Chem., 238, 2022
|
|
7DGB
 
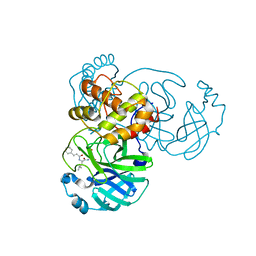 | |
7DGH
 
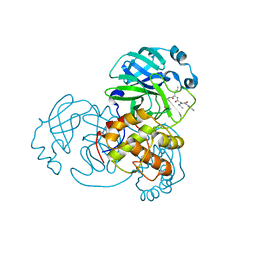 | |
7DGF
 
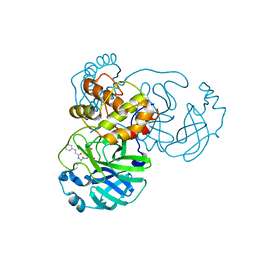 | |
7DGG
 
 | |
7DGI
 
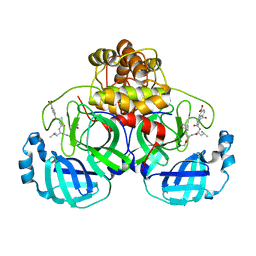 | |
7WVP
 
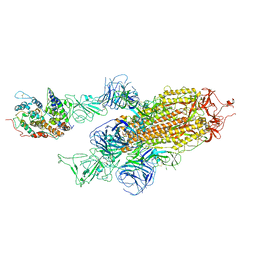 | |
7WVQ
 
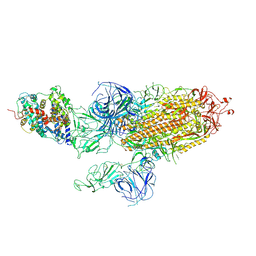 | |
8JP2
 
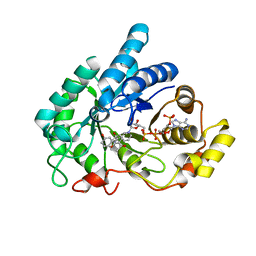 | | Crystal structure of AKR1C1 in complex with DFV | | Descriptor: | 7-HYDROXY-2-(4-HYDROXY-PHENYL)-CHROMAN-4-ONE, Aldo-keto reductase family 1 member C1, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Zheng, X.H, Liu, H, Yao, Z.Q, Zhang, L.P. | | Deposit date: | 2023-06-10 | | Release date: | 2024-04-24 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Inhibition of AKR1Cs by liquiritigenin and the structural basis.
Chem.Biol.Interact., 385, 2023
|
|
8JP1
 
 | | Crystal structure of AKR1C3 in complex with DFV | | Descriptor: | 7-HYDROXY-2-(4-HYDROXY-PHENYL)-CHROMAN-4-ONE, Aldo-keto reductase family 1 member C3, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Zheng, X.H, Liu, H, Yao, Z.Q, Zhang, L.P. | | Deposit date: | 2023-06-10 | | Release date: | 2024-04-24 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Inhibition of AKR1Cs by liquiritigenin and the structural basis.
Chem.Biol.Interact., 385, 2023
|
|
7VXC
 
 | | SARS-CoV-2 Kappa variant spike protein in C3 state | | Descriptor: | Angiotensin-converting enzyme 2, Spike glycoprotein | | Authors: | Xu, C, Cong, Y. | | Deposit date: | 2021-11-12 | | Release date: | 2021-12-22 | | Last modified: | 2022-02-16 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Conformational dynamics of the Beta and Kappa SARS-CoV-2 spike proteins and their complexes with ACE2 receptor revealed by cryo-EM.
Nat Commun, 12, 2021
|
|
