2EAV
 
 | |
6H3R
 
 | |
2XKA
 
 | |
5IL1
 
 | | Crystal structure of SAM-bound METTL3-METTL14 complex | | Descriptor: | 1,2-ETHANEDIOL, METTL14, METTL3, ... | | Authors: | Wang, X, Guan, Z, Zou, T, Yin, P. | | Deposit date: | 2016-03-04 | | Release date: | 2016-05-25 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | Structural basis of N6-adenosine methylation by the METTL3-METTL14 complex
Nature, 534, 2016
|
|
5IL0
 
 | | Crystal structural of the METTL3-METTL14 complex for N6-adenosine methylation | | Descriptor: | 1,2-ETHANEDIOL, BROMIDE ION, METTL14, ... | | Authors: | Wang, X, Guan, Z, Zou, T, Yin, P. | | Deposit date: | 2016-03-04 | | Release date: | 2016-05-25 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.882 Å) | | Cite: | Structural basis of N6-adenosine methylation by the METTL3-METTL14 complex
Nature, 534, 2016
|
|
5IL2
 
 | | Crystal structure of SAH-bound METTL3-METTL14 complex | | Descriptor: | 1,2-ETHANEDIOL, METTL14, METTL3, ... | | Authors: | Wang, X, Guan, Z, Zou, T, Yin, P. | | Deposit date: | 2016-03-04 | | Release date: | 2016-05-25 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.606 Å) | | Cite: | Structural basis of N6-adenosine methylation by the METTL3-METTL14 complex
Nature, 534, 2016
|
|
4FI9
 
 | | Structure of human SUN-KASH complex | | Descriptor: | Nesprin-2, SUN domain-containing protein 2 | | Authors: | Wang, W.J, Shi, Z.B. | | Deposit date: | 2012-06-08 | | Release date: | 2012-07-18 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (3.05 Å) | | Cite: | Structural insights into SUN-KASH complexes across the nuclear envelope.
Cell Res., 22, 2012
|
|
9M3N
 
 | | Crystal structure of human TRIM21 PRYSPRY in complex with T-02 | | Descriptor: | 1-ethyl-~{N}-methyl-5-phenyl-~{N}-[3-[3-(trifluoromethyl)phenyl]cyclobutyl]pyrazole-4-carboxamide, E3 ubiquitin-protein ligase TRIM21 | | Authors: | Zhang, L.J, Zhang, L.Y. | | Deposit date: | 2025-03-03 | | Release date: | 2025-07-16 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | TRIM21-Driven Degradation of BRD4: Development of Heterobifunctional Degraders and Investigation of Recruitment and Selectivity Mechanisms.
J.Chem.Inf.Model., 2025
|
|
5H0N
 
 | |
2JQC
 
 | | A L-amino acid mutant of a D-amino acid containing conopeptide | | Descriptor: | L-mr12 | | Authors: | Huang, F, Du, W, Han, Y, Wang, C, Chi, C. | | Deposit date: | 2007-05-31 | | Release date: | 2008-04-15 | | Last modified: | 2023-12-20 | | Method: | SOLUTION NMR | | Cite: | Purification and structural characterization of a d-amino acid-containing conopeptide, conomarphin, from Conus marmoreus
Febs J., 275, 2008
|
|
3EIJ
 
 | | Crystal structure of Pdcd4 | | Descriptor: | Programmed cell death protein 4 | | Authors: | Loh, P.G. | | Deposit date: | 2008-09-16 | | Release date: | 2009-02-17 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural basis for translational inhibition by the tumour suppressor Pdcd4
Embo J., 28, 2009
|
|
3EIQ
 
 | | Crystal structure of Pdcd4-eIF4A | | Descriptor: | Eukaryotic initiation factor 4A-I, Programmed cell death protein 4 | | Authors: | Loh, P.G, Cheng, Z, Song, H. | | Deposit date: | 2008-09-17 | | Release date: | 2009-02-24 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Structural basis for translational inhibition by the tumour suppressor Pdcd4
Embo J., 28, 2009
|
|
7YFI
 
 | | Structure of the Rat tri-heteromeric GluN1-GluN2A-GluN2C NMDA receptor in complex with glycine and glutamate | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, GLUTAMIC ACID, ... | | Authors: | Zhang, M, Zhang, J, Guo, F, Li, Y, Zhu, S. | | Deposit date: | 2022-07-08 | | Release date: | 2023-03-29 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Distinct structure and gating mechanism in diverse NMDA receptors with GluN2C and GluN2D subunits.
Nat.Struct.Mol.Biol., 30, 2023
|
|
7MFH
 
 | | Crystal structure of BIO-32546 bound mouse Autotaxin | | Descriptor: | (1R,3S,5S)-8-{(1S)-1-[8-(trifluoromethyl)-7-{[(1s,4R)-4-(trifluoromethyl)cyclohexyl]oxy}naphthalen-2-yl]ethyl}-8-azabicyclo[3.2.1]octane-3-carboxylic acid, 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Chodaparambil, J.V. | | Deposit date: | 2021-04-09 | | Release date: | 2022-04-13 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Discovery of Potent Selective Nonzinc Binding Autotaxin Inhibitor BIO-32546.
Acs Med.Chem.Lett., 12, 2021
|
|
7BZ5
 
 | | Structure of COVID-19 virus spike receptor-binding domain complexed with a neutralizing antibody | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of B38, Light chain of B38, ... | | Authors: | Wu, Y, Qi, J, Gao, F. | | Deposit date: | 2020-04-26 | | Release date: | 2020-05-13 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | A noncompeting pair of human neutralizing antibodies block COVID-19 virus binding to its receptor ACE2.
Science, 368, 2020
|
|
7YFG
 
 | | Structure of the Rat GluN1-GluN2C NMDA receptor in complex with glycine and glutamate (major class in asymmetry) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, GLUTAMIC ACID, ... | | Authors: | Zhang, M, Zhang, J, Guo, F, Li, Y, Zhu, S. | | Deposit date: | 2022-07-08 | | Release date: | 2023-03-29 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Distinct structure and gating mechanism in diverse NMDA receptors with GluN2C and GluN2D subunits.
Nat.Struct.Mol.Biol., 30, 2023
|
|
7YFM
 
 | | Structure of GluN1b-GluN2D NMDA receptor in complex with agonists glycine and glutamate. | | Descriptor: | Glutamate receptor ionotropic, NMDA 2D, Isoform 6 of Glutamate receptor ionotropic, ... | | Authors: | Zhang, J.L, Zhu, S.J, Zhang, M. | | Deposit date: | 2022-07-08 | | Release date: | 2023-03-29 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (5.1 Å) | | Cite: | Distinct structure and gating mechanism in diverse NMDA receptors with GluN2C and GluN2D subunits.
Nat.Struct.Mol.Biol., 30, 2023
|
|
7YFH
 
 | | Structure of the Rat GluN1-GluN2C NMDA receptor in complex with glycine, glutamate and (R)-PYD-106 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, GLUTAMIC ACID, ... | | Authors: | Zhang, M, Zhang, J, Guo, F, Li, Y, Zhu, S. | | Deposit date: | 2022-07-08 | | Release date: | 2023-03-29 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Distinct structure and gating mechanism in diverse NMDA receptors with GluN2C and GluN2D subunits.
Nat.Struct.Mol.Biol., 30, 2023
|
|
7YFF
 
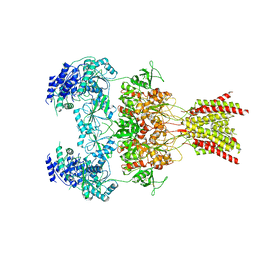 | | Structure of GluN1a-GluN2D NMDA receptor in complex with agonist glycine and competitive antagonist CPP. | | Descriptor: | (2R)-4-(3-phosphonopropyl)piperazine-2-carboxylic acid, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Zhang, J.L, Zhu, S.J, Zhang, M. | | Deposit date: | 2022-07-08 | | Release date: | 2023-04-12 | | Last modified: | 2023-08-02 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Distinct structure and gating mechanism in diverse NMDA receptors with GluN2C and GluN2D subunits.
Nat.Struct.Mol.Biol., 30, 2023
|
|
7YFL
 
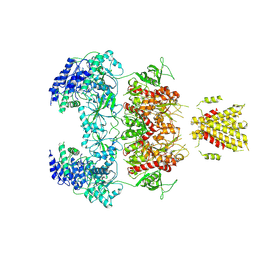 | | Structure of GluN1a-GluN2D NMDA receptor in complex with agonists glycine and glutamate. | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, GLUTAMIC ACID, ... | | Authors: | Zhang, J.L, Zhu, S.J, Zhang, M. | | Deposit date: | 2022-07-08 | | Release date: | 2023-04-12 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Distinct structure and gating mechanism in diverse NMDA receptors with GluN2C and GluN2D subunits.
Nat.Struct.Mol.Biol., 30, 2023
|
|
7YFO
 
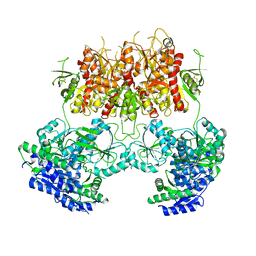 | |
7YFR
 
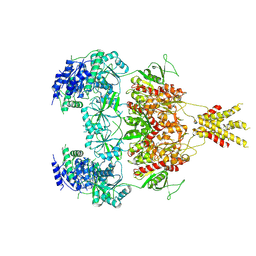 | | Structure of GluN1a E698C-GluN2D NMDA receptor in cystines non-crosslinked state. | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, GLUTAMIC ACID, ... | | Authors: | Zhang, J.L, Zhu, S.J, Zhang, M. | | Deposit date: | 2022-07-09 | | Release date: | 2023-04-12 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (5.1 Å) | | Cite: | Distinct structure and gating mechanism in diverse NMDA receptors with GluN2C and GluN2D subunits.
Nat.Struct.Mol.Biol., 30, 2023
|
|
9JM0
 
 | | retron Ec86-effector fiber | | Descriptor: | DNA (85-MER), NICOTINAMIDE, RNA (5'-R(P*CP*GP*UP*AP*AP*GP*GP*GP*UP*GP*CP*GP*CP*A)-3'), ... | | Authors: | Wang, Y.J, Guan, Z.Y, Wang, C, Zou, T.T. | | Deposit date: | 2024-09-20 | | Release date: | 2024-12-18 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | DNA methylation activates retron Ec86 filaments for antiphage defense.
Cell Rep, 43, 2024
|
|
5ISY
 
 | | Crystal structure of Nudix family protein with NAD | | Descriptor: | NADH pyrophosphatase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ZINC ION | | Authors: | Zhang, D, Guan, Z, Zou, T, Yin, P. | | Deposit date: | 2016-03-15 | | Release date: | 2016-09-14 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.354 Å) | | Cite: | Structural basis of prokaryotic NAD-RNA decapping by NudC
Cell Res., 26, 2016
|
|
4TVZ
 
 | | Crystal Structure of SCARB2 in Neural Condition (pH7.5) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Scavenger receptor class B member 2, ... | | Authors: | Dang, M.H, Wang, X.X, Rao, Z.H. | | Deposit date: | 2014-06-29 | | Release date: | 2015-07-08 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.006 Å) | | Cite: | Molecular mechanism of SCARB2-mediated attachment and uncoating of EV71
Protein Cell, 5, 2014
|
|
