7R7N
 
 | |
7R6X
 
 | | SARS-CoV-2 spike receptor-binding domain (RBD) in complex with S2E12 Fab, S309 Fab, and S304 Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, Monoclonal antibody S2E12 Fab heavy chain, ... | | Authors: | Snell, G, Czudnochowski, N, Croll, T.I, Nix, J.C, Corti, D, Cameroni, E, Pinto, D, Beltramello, M. | | Deposit date: | 2021-06-23 | | Release date: | 2021-07-21 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | SARS-CoV-2 RBD antibodies that maximize breadth and resistance to escape.
Nature, 597, 2021
|
|
7R6W
 
 | | SARS-CoV-2 spike receptor-binding domain (RBD) in complex with S2X35 Fab and S309 Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, GLYCEROL, ... | | Authors: | Snell, G, Czudnochowski, N, Hernandez, P, Nix, J.C, Croll, T.I, Corti, D, Cameroni, E, Pinto, D, Beltramello, M. | | Deposit date: | 2021-06-23 | | Release date: | 2021-07-21 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | SARS-CoV-2 RBD antibodies that maximize breadth and resistance to escape.
Nature, 597, 2021
|
|
7Y5A
 
 | | Cryo-EM structure of the Mycolicibacterium smegmatis F1-ATPase | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ATP synthase gamma chain, ... | | Authors: | Wong, C.F, Saw, W.-G, Grueber, G. | | Deposit date: | 2022-06-16 | | Release date: | 2022-11-23 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structural Elements Involved in ATP Hydrolysis Inhibition and ATP Synthesis of Tuberculosis and Nontuberculous Mycobacterial F-ATP Synthase Decipher New Targets for Inhibitors.
Antimicrob.Agents Chemother., 66, 2022
|
|
7Y5C
 
 | | Cryo-EM structure of F-ATP synthase from Mycolicibacterium smegmatis (rotational state 2) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ATP synthase epsilon chain, ... | | Authors: | Saw, W.-G, Wong, C.F, Grueber, G. | | Deposit date: | 2022-06-16 | | Release date: | 2022-11-23 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (4.7 Å) | | Cite: | Structural Elements Involved in ATP Hydrolysis Inhibition and ATP Synthesis of Tuberculosis and Nontuberculous Mycobacterial F-ATP Synthase Decipher New Targets for Inhibitors.
Antimicrob.Agents Chemother., 66, 2022
|
|
7Y5B
 
 | | Cryo-EM structure of F-ATP synthase from Mycolicibacterium smegmatis (rotational state 1) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ATP synthase epsilon chain, ... | | Authors: | Saw, W.-G, Wong, C.F, Grueber, G. | | Deposit date: | 2022-06-16 | | Release date: | 2022-11-23 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (4.4 Å) | | Cite: | Structural Elements Involved in ATP Hydrolysis Inhibition and ATP Synthesis of Tuberculosis and Nontuberculous Mycobacterial F-ATP Synthase Decipher New Targets for Inhibitors.
Antimicrob.Agents Chemother., 66, 2022
|
|
7Y5D
 
 | | Cryo-EM structure of F-ATP synthase from Mycolicibacterium smegmatis (rotational state 3) (backbone) | | Descriptor: | ATP synthase epsilon chain, ATP synthase gamma chain, ATP synthase subunit a, ... | | Authors: | Saw, W.-G, Wong, C.F, Grueber, G. | | Deposit date: | 2022-06-16 | | Release date: | 2022-11-23 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (7.3 Å) | | Cite: | Structural Elements Involved in ATP Hydrolysis Inhibition and ATP Synthesis of Tuberculosis and Nontuberculous Mycobacterial F-ATP Synthase Decipher New Targets for Inhibitors.
Antimicrob.Agents Chemother., 66, 2022
|
|
8GER
 
 | | E. eligens beta-glucuronidase bound to norquetiapine-glucuronide | | Descriptor: | 11-(4-beta-D-glucopyranuronosylpiperazin-1-yl)dibenzo[b,f][1,4]thiazepine, Beta-glucuronidase | | Authors: | Simpson, J.B, Lietzan, A.D, Redinbo, M.R. | | Deposit date: | 2023-03-07 | | Release date: | 2024-03-20 | | Last modified: | 2024-06-26 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Gut microbial beta-glucuronidases influence endobiotic homeostasis and are modulated by diverse therapeutics.
Cell Host Microbe, 32, 2024
|
|
8GEO
 
 | | E. eligens beta-glucuronidase bound to 3-OH-desloratidine-glucuronide | | Descriptor: | 8-chloro-11-(1-beta-D-glucopyranuronosylpiperidin-4-ylidene)-3-hydroxy-6,11-dihydro-5H-benzo[5,6]cyclohepta[1,2-b]pyridine, Beta-glucuronidase, GLYCEROL | | Authors: | Simpson, J.B, Redinbo, M.R. | | Deposit date: | 2023-03-07 | | Release date: | 2024-03-20 | | Last modified: | 2024-06-26 | | Method: | X-RAY DIFFRACTION (2.89 Å) | | Cite: | Gut microbial beta-glucuronidases influence endobiotic homeostasis and are modulated by diverse therapeutics.
Cell Host Microbe, 32, 2024
|
|
8GET
 
 | | R. hominis 2 beta-glucuronidase bound to norquetiapine-glucuronide | | Descriptor: | 11-(4-beta-D-glucopyranuronosylpiperazin-1-yl)dibenzo[b,f][1,4]thiazepine, FLAVIN MONONUCLEOTIDE, GLYCEROL, ... | | Authors: | Simpson, J.B, Redinbo, M.R. | | Deposit date: | 2023-03-07 | | Release date: | 2024-03-20 | | Last modified: | 2024-06-26 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Gut microbial beta-glucuronidases influence endobiotic homeostasis and are modulated by diverse therapeutics.
Cell Host Microbe, 32, 2024
|
|
8E72
 
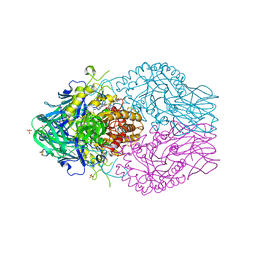 | | Treponema lecithinolyticum beta-glucuronidase in complex with a ciprofloxacin-glucuronide conjugate | | Descriptor: | 3-carboxy-1-cyclopropyl-6-fluoro-7-(4-beta-D-glucopyranuronosyl-3,4-dihydropyrazin-1(2H)-yl)-4-oxo-1,4-dihydroquinoline, Glycosyl hydrolase family 2, TIM barrel domain protein, ... | | Authors: | Lietzan, A.D, Redinbo, M.R. | | Deposit date: | 2022-08-23 | | Release date: | 2023-06-21 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Microbial beta-glucuronidases drive human periodontal disease etiology.
Sci Adv, 9, 2023
|
|
8HGX
 
 | |
3SXF
 
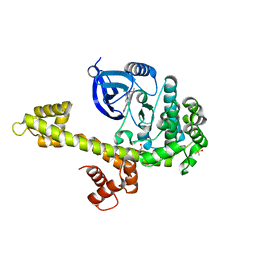 | | Calcium-Dependent Protein Kinase 1 from Toxoplasma gondii (TgCDPK1) in complex with Bumped Kinase Inhibitor, RM-1-89 | | Descriptor: | 1,2-ETHANEDIOL, 3-(6-ethoxynaphthalen-2-yl)-1-(propan-2-yl)-1H-pyrazolo[3,4-d]pyrimidin-4-amine, Calmodulin-domain protein kinase 1, ... | | Authors: | Larson, E.T, Merritt, E.A. | | Deposit date: | 2011-07-14 | | Release date: | 2012-03-14 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Multiple determinants for selective inhibition of apicomplexan calcium-dependent protein kinase CDPK1.
J.Med.Chem., 55, 2012
|
|
3T3V
 
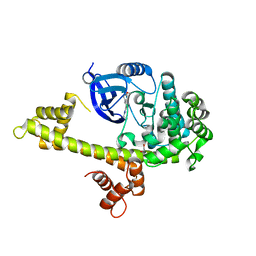 | | Calcium-Dependent Protein Kinase 1 from Toxoplasma gondii (TgCDPK1) in complex with Bumped Kinase Inhibitor, RM-1-87 | | Descriptor: | 1,2-ETHANEDIOL, 3-(6-methoxynaphthalen-2-yl)-1-(propan-2-yl)-1H-pyrazolo[3,4-d]pyrimidin-4-amine, Calmodulin-domain protein kinase 1 | | Authors: | Larson, E.T, Merritt, E.A. | | Deposit date: | 2011-07-25 | | Release date: | 2012-03-14 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Multiple determinants for selective inhibition of apicomplexan calcium-dependent protein kinase CDPK1.
J.Med.Chem., 55, 2012
|
|
3T3U
 
 | | Calcium-Dependent Protein Kinase 1 from Toxoplasma gondii (TgCDPK1) in complex with Bumped Kinase Inhibitor, RM-1-130 | | Descriptor: | 1,2-ETHANEDIOL, 3-(6-methoxynaphthalen-2-yl)-1-(piperidin-4-ylmethyl)-1H-pyrazolo[3,4-d]pyrimidin-4-amine, Calmodulin-domain protein kinase 1, ... | | Authors: | Larson, E.T, Merritt, E.A. | | Deposit date: | 2011-07-25 | | Release date: | 2012-03-14 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Multiple determinants for selective inhibition of apicomplexan calcium-dependent protein kinase CDPK1.
J.Med.Chem., 55, 2012
|
|
1NNC
 
 | |
4M84
 
 | |
1PBF
 
 | | CRYSTAL STRUCTURES OF WILD-TYPE P-HYDROXYBENZOATE HYDROXYLASE COMPLEXED WITH 4-AMINOBENZOATE, 2,4-DIHYDROXYBENZOATE AND 2-HYDROXY-4-AMINOBENZOATE AND OF THE TRY222ALA MUTANT, COMPLEXED WITH 2-HYDROXY-4-AMINOBENZOATE. EVIDENCE FOR A PROTON CHANNEL AND A NEW BINDING MODE OF THE FLAVIN RING | | Descriptor: | 2-HYDROXY-4-AMINOBENZOIC ACID, FLAVIN-ADENINE DINUCLEOTIDE, P-HYDROXYBENZOATE HYDROXYLASE | | Authors: | Schreuder, H.A, Van Der Bolt, F.J.T, Van Berkel, W.J.H. | | Deposit date: | 1994-07-06 | | Release date: | 1994-09-30 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structures of wild-type p-hydroxybenzoate hydroxylase complexed with 4-aminobenzoate,2,4-dihydroxybenzoate, and 2-hydroxy-4-aminobenzoate and of the Tyr222Ala mutant complexed with 2-hydroxy-4-aminobenzoate. Evidence for a proton channel and a new binding mode of the flavin ring
Biochemistry, 33, 1994
|
|
1PBC
 
 | | CRYSTAL STRUCTURES OF WILD-TYPE P-HYDROXYBENZOATE HYDROXYLASE COMPLEXED WITH 4-AMINOBENZOATE, 2,4-DIHYDROXYBENZOATE AND 2-HYDROXY-4-AMINOBENZOATE AND OF THE TRY222ALA MUTANT, COMPLEXED WITH 2-HYDROXY-4-AMINOBENZOATE. EVIDENCE FOR A PROTON CHANNEL AND A NEW BINDING MODE OF THE FLAVIN RING | | Descriptor: | 2-HYDROXY-4-AMINOBENZOIC ACID, FLAVIN-ADENINE DINUCLEOTIDE, P-HYDROXYBENZOATE HYDROXYLASE | | Authors: | Schreuder, H.A, Van Der Bolt, F.J.T, Van Berkel, W.J.H. | | Deposit date: | 1994-07-06 | | Release date: | 1994-09-30 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structures of wild-type p-hydroxybenzoate hydroxylase complexed with 4-aminobenzoate,2,4-dihydroxybenzoate, and 2-hydroxy-4-aminobenzoate and of the Tyr222Ala mutant complexed with 2-hydroxy-4-aminobenzoate. Evidence for a proton channel and a new binding mode of the flavin ring
Biochemistry, 33, 1994
|
|
5VQW
 
 | | Crystal Structure of HIV-1 Reverse Transcriptase (Y181C) Variant in Complex with N-(6-cyano-3-(2-(2-(2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl)ethoxy)phenoxy)-4-methylnaphthalen-1-yl)acrylamide (JLJ685), a Non-nucleoside Inhibitor | | Descriptor: | N-(6-cyano-3-{2-[2-(2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl)ethoxy]phenoxy}-4-methylnaphthalen-1-yl)prop-2-enamide, Reverse transcriptase/ribonuclease H, SULFATE ION, ... | | Authors: | Chan, A.H, Kudalkar, S.N, Anderson, K.S. | | Deposit date: | 2017-05-09 | | Release date: | 2017-08-23 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Covalent inhibitors for eradication of drug-resistant HIV-1 reverse transcriptase: From design to protein crystallography.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
5VQR
 
 | | Crystal Structure of HIV-1 Reverse Transcriptase in Complex with N-(6-cyano-3-(2-(2-(2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl)ethoxy)phenoxy)-4-methylnaphthalen-1-yl)-N-methylacrylamide (JLJ684), a Non-nucleoside Inhibitor | | Descriptor: | N-(6-cyano-3-{2-[2-(2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl)ethoxy]phenoxy}-4-methylnaphthalen-1-yl)-N-methylprop-2-enamide, Reverse transcriptase/ribonuclease H, SULFATE ION, ... | | Authors: | Chan, A.H, Anderson, K.S. | | Deposit date: | 2017-05-09 | | Release date: | 2017-08-23 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Covalent inhibitors for eradication of drug-resistant HIV-1 reverse transcriptase: From design to protein crystallography.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
5VQZ
 
 | | Crystal Structure of HIV-1 Reverse Transcriptase (K103N, Y181C) Variant in Complex with 2-chloro-N-(6-cyano-3-(2-(2-(2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl)ethoxy)phenoxy)-4-methylnaphthalen-1-yl)-N-methylacetamide (JLJ686), a Non-nucleoside Inhibitor | | Descriptor: | N-(6-cyano-3-{2-[2-(2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl)ethoxy]phenoxy}-4-methylnaphthalen-1-yl)-N-methylacetamide, Reverse transcriptase/ribonuclease H, SULFATE ION, ... | | Authors: | Buckingham, A.B, Chan, A.H, Anderson, K.S. | | Deposit date: | 2017-05-09 | | Release date: | 2017-08-23 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | Covalent inhibitors for eradication of drug-resistant HIV-1 reverse transcriptase: From design to protein crystallography.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
5VQX
 
 | | Crystal Structure of HIV-1 Reverse Transcriptase (Y181C) Variant in Complex with 2-chloro-N-(6-cyano-3-(2-(2-(2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl)ethoxy)phenoxy)-4-methylnaphthalen-1-yl)-N-methylacetamide (JLJ686), a Non-nucleoside Inhibitor | | Descriptor: | N-(6-cyano-3-{2-[2-(2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl)ethoxy]phenoxy}-4-methylnaphthalen-1-yl)-N-methylacetamide, Reverse transcriptase/ribonuclease H, SULFATE ION, ... | | Authors: | Chan, A.H, Anderson, K.S. | | Deposit date: | 2017-05-09 | | Release date: | 2017-08-23 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Covalent inhibitors for eradication of drug-resistant HIV-1 reverse transcriptase: From design to protein crystallography.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
5VQS
 
 | | Crystal Structure of HIV-1 Reverse Transcriptase in Complex with N-(6-cyano-3-(2-(2-(2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl)ethoxy)phenoxy)-4-methylnaphthalen-1-yl)acrylamide (JLJ685), a Non-nucleoside Inhibitor | | Descriptor: | N-(6-cyano-3-{2-[2-(2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl)ethoxy]phenoxy}-4-methylnaphthalen-1-yl)prop-2-enamide, Reverse transcriptase/ribonuclease H, SULFATE ION, ... | | Authors: | Chan, A.H, Anderson, K.S. | | Deposit date: | 2017-05-09 | | Release date: | 2017-08-23 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.504 Å) | | Cite: | Covalent inhibitors for eradication of drug-resistant HIV-1 reverse transcriptase: From design to protein crystallography.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
5FU4
 
 | | The complexity of the Ruminococcus flavefaciens cellulosome reflects an expansion in glycan recognition | | Descriptor: | CBM74-RFGH5, beta-D-mannopyranose-(1-4)-beta-D-mannopyranose-(1-4)-beta-D-mannopyranose, beta-D-mannopyranose-(1-4)-beta-D-mannopyranose-(1-4)-beta-D-mannopyranose-(1-4)-beta-D-mannopyranose-(1-4)-beta-D-mannopyranose | | Authors: | Basle, A, Luis, A.S, Venditto, I, Gilbert, H.J. | | Deposit date: | 2016-01-20 | | Release date: | 2016-06-22 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Complexity of the Ruminococcus flavefaciens cellulosome reflects an expansion in glycan recognition.
Proc. Natl. Acad. Sci. U.S.A., 113, 2016
|
|
