22GS
 
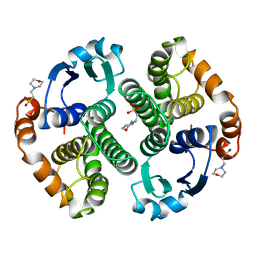 | | HUMAN GLUTATHIONE S-TRANSFERASE P1-1 Y49F MUTANT | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, GLUTATHIONE S-TRANSFERASE P1-1 | | Authors: | Oakley, A.J. | | Deposit date: | 1998-03-10 | | Release date: | 1999-03-23 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Thermodynamic description of the effect of the mutation Y49F on human glutathione transferase P1-1 in binding with glutathione and the inhibitor S-hexylglutathione.
J.Biol.Chem., 278, 2003
|
|
8GSS
 
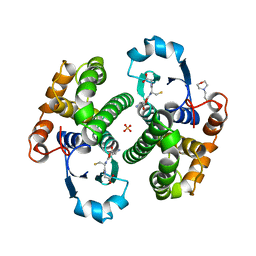 | | Human glutathione S-transferase P1-1, complex with glutathione | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, GLUTATHIONE, GLUTATHIONE S-TRANSFERASE P1-1, ... | | Authors: | Oakley, A, Parker, M. | | Deposit date: | 1997-08-14 | | Release date: | 1998-09-16 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The structures of human glutathione transferase P1-1 in complex with glutathione and various inhibitors at high resolution.
J.Mol.Biol., 274, 1997
|
|
10GS
 
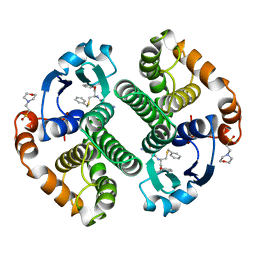 | | HUMAN GLUTATHIONE S-TRANSFERASE P1-1, COMPLEX WITH TER117 | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, GLUTATHIONE S-TRANSFERASE P1-1, L-gamma-glutamyl-S-benzyl-N-[(S)-carboxy(phenyl)methyl]-L-cysteinamide | | Authors: | Oakley, A, Parker, M. | | Deposit date: | 1997-08-14 | | Release date: | 1998-09-16 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The structures of human glutathione transferase P1-1 in complex with glutathione and various inhibitors at high resolution.
J.Mol.Biol., 274, 1997
|
|
5T3N
 
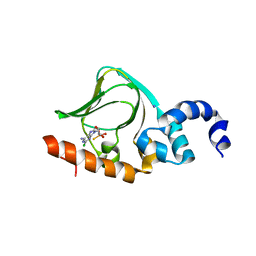 | | Sp-2Cl-cAMPS bound to PKAR CBD2 | | Descriptor: | (2S,4aR,6R,7R,7aS)-6-(6-amino-2-chloro-9H-purin-9-yl)-7-hydroxy-2-sulfanyltetrahydro-2H,4H-2lambda~5~-furo[3,2-d][1,3,2]dioxaphosphinin-2-one, IODIDE ION, cAMP-dependent protein kinase regulatory subunit | | Authors: | Littler, D.R, Gilson, P. | | Deposit date: | 2016-08-26 | | Release date: | 2016-10-12 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Disrupting the Allosteric Interaction between the Plasmodium falciparum cAMP-dependent Kinase and Its Regulatory Subunit.
J. Biol. Chem., 291, 2016
|
|
6XQA
 
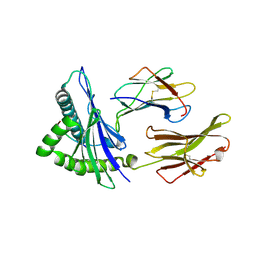 | | Crystal Structure of HLA A*2402 in complex with TYQWVLKNL, an 9-mer epitope from Influenza B virus | | Descriptor: | Beta-2-microglobulin, MAGNESIUM ION, MHC class I antigen, ... | | Authors: | Nguyen, A.T, Szeto, C, Gras, S. | | Deposit date: | 2020-07-09 | | Release date: | 2021-04-14 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.16 Å) | | Cite: | CD8 + T cell landscape in Indigenous and non-Indigenous people restricted by influenza mortality-associated HLA-A*24:02 allomorph.
Nat Commun, 12, 2021
|
|
2DUT
 
 | |
5GSS
 
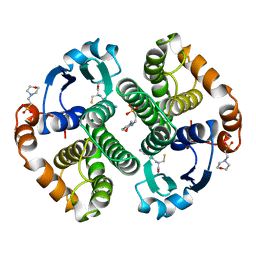 | | HUMAN GLUTATHIONE S-TRANSFERASE P1-1, COMPLEX WITH GLUTATHIONE | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, GLUTATHIONE, GLUTATHIONE S-TRANSFERASE P1-1 | | Authors: | Oakley, A, Parker, M. | | Deposit date: | 1997-08-11 | | Release date: | 1998-09-16 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | The structures of human glutathione transferase P1-1 in complex with glutathione and various inhibitors at high resolution.
J.Mol.Biol., 274, 1997
|
|
2EYT
 
 | |
2G3O
 
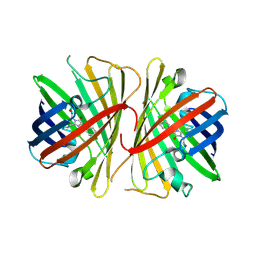 | | The 2.1A crystal structure of copGFP | | Descriptor: | green fluorescent protein 2 | | Authors: | Wilmann, P.G. | | Deposit date: | 2006-02-20 | | Release date: | 2006-08-15 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The 2.1A crystal structure of copGFP, a representative member of the copepod clade within the green fluorescent protein superfamily
J.Mol.Biol., 359, 2006
|
|
7S8R
 
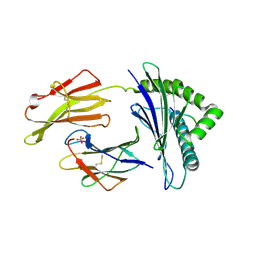 | | Crystal Structure of HLA A*1101 in complex with SALEWIKNK, an 9-mer epitope from Influenza B | | Descriptor: | Beta-2-microglobulin, HLA class I histocompatibility antigen, A alpha chain, ... | | Authors: | Nguyen, A.T, Szeto, C, Gras, S. | | Deposit date: | 2021-09-19 | | Release date: | 2022-02-23 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | HLA-A*11:01-restricted CD8+ T cell immunity against influenza A and influenza B viruses in Indigenous and non-Indigenous people.
Plos Pathog., 18, 2022
|
|
7S8S
 
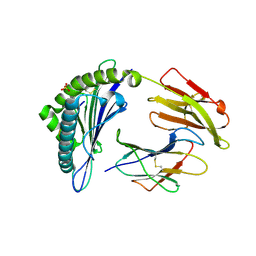 | | Crystal Structure of HLA A*1101 in complex with RVLVNGTFLK, an 10-mer epitope from Influenza B | | Descriptor: | Beta-2-microglobulin, HLA class I histocompatibility antigen, A alpha chain, ... | | Authors: | Nguyen, A.T, Szeto, C, Gras, S. | | Deposit date: | 2021-09-19 | | Release date: | 2022-02-23 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | HLA-A*11:01-restricted CD8+ T cell immunity against influenza A and influenza B viruses in Indigenous and non-Indigenous people.
Plos Pathog., 18, 2022
|
|
7S8Q
 
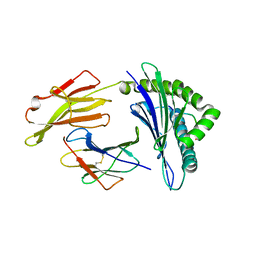 | | Crystal Structure of HLA A*1101 in complex with KTNGNAFIGK, an 10-mer epitope from Influenza B | | Descriptor: | Beta-2-microglobulin, HLA class I histocompatibility antigen, A alpha chain, ... | | Authors: | Nguyen, A.T, Szeto, C, Gras, S. | | Deposit date: | 2021-09-19 | | Release date: | 2022-02-23 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | HLA-A*11:01-restricted CD8+ T cell immunity against influenza A and influenza B viruses in Indigenous and non-Indigenous people.
Plos Pathog., 18, 2022
|
|
6NF7
 
 | | Crystal Structure of RT1.Aa-Bu31-10 | | Descriptor: | Beta-2-microglobulin, Bu31-10 peptide, RT1A.a | | Authors: | Gras, S. | | Deposit date: | 2018-12-19 | | Release date: | 2019-12-25 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Cross-Reactive Donor-Specific CD8+Tregs Efficiently Prevent Transplant Rejection.
Cell Rep, 29, 2019
|
|
2PO6
 
 | |
2EYR
 
 | |
6PBH
 
 | |
2H4P
 
 | |
2OKK
 
 | | The X-ray crystal structure of the 65kDa isoform of Glutamic Acid Decarboxylase (GAD65) | | Descriptor: | GAMMA-AMINO-BUTANOIC ACID, GLYCEROL, Glutamate decarboxylase 2 | | Authors: | Buckle, A.M, Fenalti, G, Law, R.H.P, Whisstock, J.C. | | Deposit date: | 2007-01-17 | | Release date: | 2007-03-27 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | GABA production by glutamic acid decarboxylase is regulated by a dynamic catalytic loop.
Nat.Struct.Mol.Biol., 14, 2007
|
|
2FYY
 
 | |
2FZ3
 
 | |
2OKJ
 
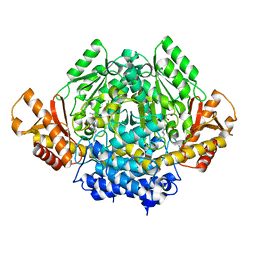 | | The X-ray crystal structure of the 67kDa isoform of Glutamic Acid Decarboxylase (GAD67) | | Descriptor: | 4-[({3-HYDROXY-2-METHYL-5-[(PHOSPHONOOXY)METHYL]PYRIDIN-4-YL}METHYL)AMINO]BUTANOIC ACID, GAMMA-AMINO-BUTANOIC ACID, Glutamate decarboxylase 1 | | Authors: | Buckle, A.M, Fenalti, G, Law, R.H.P, Whisstock, J.C. | | Deposit date: | 2007-01-17 | | Release date: | 2007-03-27 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | GABA production by glutamic acid decarboxylase is regulated by a dynamic catalytic loop.
Nat.Struct.Mol.Biol., 14, 2007
|
|
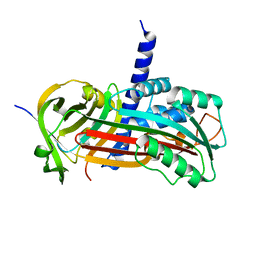
2H4Q
 
 | |
2EYS
 
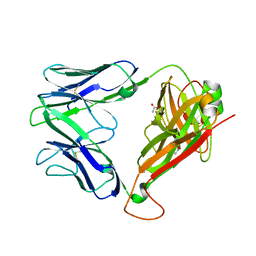 | |
3BDW
 
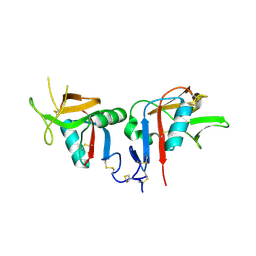 | | Human CD94/NKG2A | | Descriptor: | NKG2-A/NKG2-B type II integral membrane protein, Natural killer cells antigen CD94 | | Authors: | Sullivan, L.C, Clements, C.S. | | Deposit date: | 2007-11-15 | | Release date: | 2008-01-01 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The Heterodimeric Assembly of the CD94-NKG2 Receptor Family and Implications for Human Leukocyte Antigen-E Recognition
Immunity, 27, 2007
|
|
2H4R
 
 | |
