4KYA
 
 | | Crystal structure of non-classical TS inhibitor 3 in complex with Toxoplasma gondii TS-DHFR | | Descriptor: | 2'-DEOXYURIDINE 5'-MONOPHOSPHATE, 2-Amino-5-(1-naphthylsulfanyl)-3,9-dihydro-4H-pyrimido[4,5-b]indol-4-one, Bifunctional dihydrofolate reductase-thymidylate synthase, ... | | Authors: | Sharma, H, Anderson, K.S. | | Deposit date: | 2013-05-28 | | Release date: | 2014-08-06 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.263 Å) | | Cite: | Discovery of potent and selective inhibitors of Toxoplasma gondii thymidylate synthase for opportunistic infections.
ACS Med Chem Lett, 4, 2013
|
|
7Y6D
 
 | |
7SOT
 
 | |
7SOV
 
 | |
7SOO
 
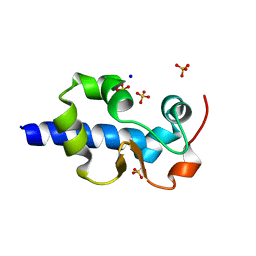 | | LaM domain of human LARP1 | | Descriptor: | Isoform 2 of La-related protein 1, SODIUM ION, SULFATE ION | | Authors: | Kozlov, G, Gehring, K. | | Deposit date: | 2021-11-01 | | Release date: | 2022-08-03 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structural basis of 3'-end poly(A) RNA recognition by LARP1.
Nucleic Acids Res., 50, 2022
|
|
7SOQ
 
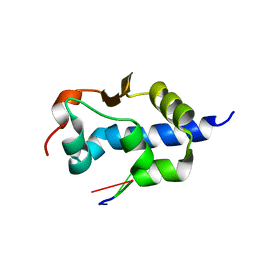 | | LaM domain of human LARP1 in complex with AAA RNA | | Descriptor: | Isoform 2 of La-related protein 1, RNA (5'-R(*AP*AP*A)-3') | | Authors: | Kozlov, G, Gehring, K. | | Deposit date: | 2021-11-01 | | Release date: | 2022-08-03 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Structural basis of 3'-end poly(A) RNA recognition by LARP1.
Nucleic Acids Res., 50, 2022
|
|
7SOR
 
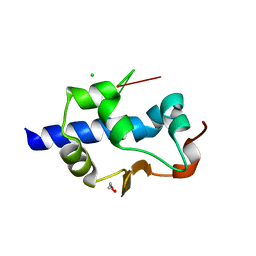 | | LaM domain of human LARP1 in complex with AAA RNA | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Isoform 2 of La-related protein 1, ... | | Authors: | Kozlov, G, Gehring, K. | | Deposit date: | 2021-11-01 | | Release date: | 2022-08-03 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Structural basis of 3'-end poly(A) RNA recognition by LARP1.
Nucleic Acids Res., 50, 2022
|
|
7SOP
 
 | |
7SOU
 
 | |
5EMY
 
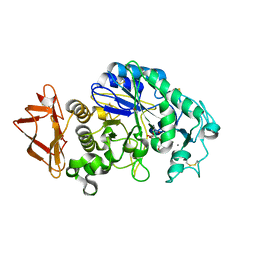 | | Human Pancreatic Alpha-Amylase in complex with the mechanism based inactivator glucosyl epi-cyclophellitol | | Descriptor: | (1R,2R,3S,5R,6S)-2,3,5-trihydroxy-6-(hydroxymethyl)cyclohexyl alpha-D-glucopyranoside, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Caner, S, Brayer, G.D. | | Deposit date: | 2015-11-06 | | Release date: | 2016-07-06 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.231 Å) | | Cite: | Glucosyl epi-cyclophellitol allows mechanism-based inactivation and structural analysis of human pancreatic alpha-amylase.
Febs Lett., 590, 2016
|
|
6VAB
 
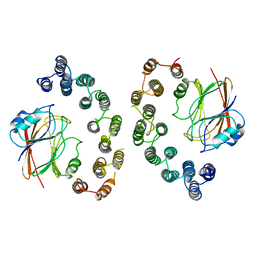 | | Mouse retromer sub-structure: VPS35/VPS35 flat dimer | | Descriptor: | Vacuolar protein sorting-associated protein 29, Vacuolar protein sorting-associated protein 35 | | Authors: | Kendall, A.K, Jackson, L.P. | | Deposit date: | 2019-12-17 | | Release date: | 2020-02-19 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (4.9 Å) | | Cite: | Mammalian Retromer Is an Adaptable Scaffold for Cargo Sorting from Endosomes.
Structure, 28, 2020
|
|
6BB1
 
 | | Lactate Dehydrogenase in complex with inhibitor (R)-5-((2-chlorophenyl)thio)-6'-(4-fluorophenoxy)-4-hydroxy-2-(thiophen-3-yl)-2,3-dihydro-[2,2'-bipyridin]-6(1H)-one | | Descriptor: | (2R)-5-[(2-chlorophenyl)sulfanyl]-6'-(4-fluorophenoxy)-4-hydroxy-2-(thiophen-3-yl)-2,3-dihydro[2,2'-bipyridin]-6(1H)-one, L-lactate dehydrogenase A chain, LACTIC ACID, ... | | Authors: | Ultsch, M, Eigenbrot, C. | | Deposit date: | 2017-10-16 | | Release date: | 2018-10-24 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure-guided optimization and in vivo activities of hydroxylactone and hydroxylactam Inhibitors of Human Lactate Dehydrogenase
To Be Published
|
|
6K8K
 
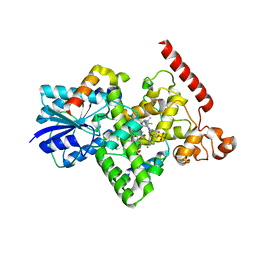 | | Crystal structure of Arabidopsis thaliana BIC2-CRY2 complex | | Descriptor: | ADENOSINE MONOPHOSPHATE, Cryptochrome-2, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Wang, X, Ma, L, Guan, Z, Yin, P. | | Deposit date: | 2019-06-12 | | Release date: | 2020-05-13 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural insights into BIC-mediated inactivation of Arabidopsis cryptochrome 2.
Nat.Struct.Mol.Biol., 27, 2020
|
|
1XWN
 
 | | solution structure of cyclophilin like 1(PPIL1) and insights into its interaction with SKIP | | Descriptor: | Peptidyl-prolyl cis-trans isomerase like 1 | | Authors: | Xu, C, Xu, Y, Tang, Y, Wu, J, Shi, Y, Huang, Q, Zhang, Q. | | Deposit date: | 2004-11-01 | | Release date: | 2005-10-18 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of human peptidyl prolyl isomerase like protein 1 and insights into its interaction with SKIP
J.Biol.Chem., 281, 2006
|
|
9BFL
 
 | |
6AUD
 
 | | PI3K-gamma K802T in complex with Cpd 8 10-((1-(tert-butyl)piperidin-4-yl)sulfinyl)-2-(1-isopropyl-1H-1,2,4-triazol-5-yl)-5,6-dihydrobenzo[f]imidazo[1,2-d][1,4]oxazepine | | Descriptor: | 10-[(S)-(1-tert-butylpiperidin-4-yl)sulfinyl]-2-[1-(propan-2-yl)-1H-1,2,4-triazol-5-yl]-5,6-dihydroimidazo[1,2-d][1,4]benzoxazepine, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit gamma isoform | | Authors: | Murray, J.M, Ultsch, M. | | Deposit date: | 2017-08-31 | | Release date: | 2017-11-15 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.015 Å) | | Cite: | Design of Selective Benzoxazepin PI3K delta Inhibitors Through Control of Dihedral Angles.
ACS Med Chem Lett, 8, 2017
|
|
6BB2
 
 | | Lactate Dehydrogenase in complex with inhibitor (S)-5-((2-chlorophenyl)thio)-6'-(4-fluorophenoxy)-4-hydroxy-2-(thiophen-3-yl)-2,3-dihydro-[2,2'-bipyridin]-6(1H)-one | | Descriptor: | (2S)-5-[(2-chlorophenyl)sulfanyl]-6'-(4-fluorophenoxy)-4-hydroxy-2-(thiophen-3-yl)-2,3-dihydro[2,2'-bipyridin]-6(1H)-one, L-lactate dehydrogenase A chain, LACTIC ACID, ... | | Authors: | Ultsch, M, Eigenbrot, C. | | Deposit date: | 2017-10-16 | | Release date: | 2018-10-24 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.47 Å) | | Cite: | Structure-guided optimization and in vivo activities of hydroxylactone and hydroxylactam Inhibitors of Human Lactate Dehydrogenase
To Be Published
|
|
6US8
 
 | |
3O51
 
 | | Crystal structure of anthranilamide 10 bound to AuroraA | | Descriptor: | N-[4-({3-[5-fluoro-2-(methylideneamino)pyrimidin-4-yl]pyridin-2-yl}oxy)phenyl]-2-(phenylamino)benzamide, cDNA FLJ58295, highly similar to Serine/threonine-protein kinase 6 | | Authors: | Huang, X. | | Deposit date: | 2010-07-27 | | Release date: | 2010-08-18 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Discovery of a potent, selective, and orally bioavailable pyridinyl-pyrimidine phthalazine aurora kinase inhibitor.
J.Med.Chem., 53, 2010
|
|
6JQ5
 
 | | The structure of Hatchet Ribozyme | | Descriptor: | MAGNESIUM ION, RNA (82-MER) | | Authors: | Ren, A, Zheng, L. | | Deposit date: | 2019-03-29 | | Release date: | 2019-06-12 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.059 Å) | | Cite: | Hatchet ribozyme structure and implications for cleavage mechanism.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
5LOL
 
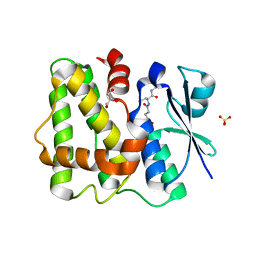 | | Glutathione-bound Dehydroascorbate Reductase 2 of Arabidopsis thaliana | | Descriptor: | GLUTATHIONE, GLYCEROL, Glutathione S-transferase DHAR2, ... | | Authors: | Young, D.R, Pallo, A, Bodra, N, Messens, J. | | Deposit date: | 2016-08-09 | | Release date: | 2017-06-07 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Arabidopsis thaliana dehydroascorbate reductase 2: Conformational flexibility during catalysis.
Sci Rep, 7, 2017
|
|
5WQC
 
 | | Crystal structure of human orexin 2 receptor bound to the selective antagonist EMPA determined by the synchrotron light source at SPring-8. | | Descriptor: | N-ethyl-2-[(6-methoxypyridin-3-yl)-(2-methylphenyl)sulfonyl-amino]-N-(pyridin-3-ylmethyl)ethanamide, OLEIC ACID, Orexin receptor type 2,GlgA glycogen synthase,Orexin receptor type 2, ... | | Authors: | Suno, R, Hirata, K, Yamashita, K, Tsujimoto, H, Sasanuma, M, Horita, S, Yamamoto, M, Rosenbaum, D.M, Iwata, S, Shimamura, T, Kobayashi, T. | | Deposit date: | 2016-11-25 | | Release date: | 2017-11-29 | | Last modified: | 2018-01-17 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Crystal Structures of Human Orexin 2 Receptor Bound to the Subtype-Selective Antagonist EMPA
Structure, 26, 2018
|
|
6V6U
 
 | |
5Y18
 
 | |
6B7H
 
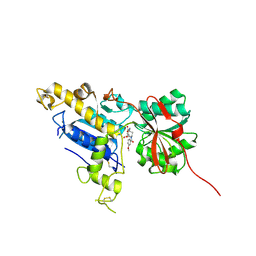 | | Structure of mGluR3 with an agonist | | Descriptor: | (1S,2S,4S,5R,6S)-2-amino-4-[(3-methoxybenzene-1-carbonyl)amino]bicyclo[3.1.0]hexane-2,6-dicarboxylic acid, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Monn, J.A, Clawson, D.K. | | Deposit date: | 2017-10-04 | | Release date: | 2018-04-25 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.82 Å) | | Cite: | Synthesis and Pharmacological Characterization of C4beta-Amide-Substituted 2-Aminobicyclo[3.1.0]hexane-2,6-dicarboxylates. Identification of (1 S,2 S,4 S,5 R,6 S)-2-Amino-4-[(3-methoxybenzoyl)amino]bicyclo[3.1.0]hexane-2,6-dicarboxylic Acid (LY2794193), a Highly Potent and Selective mGlu3Receptor Agonist.
J. Med. Chem., 61, 2018
|
|
