7VN3
 
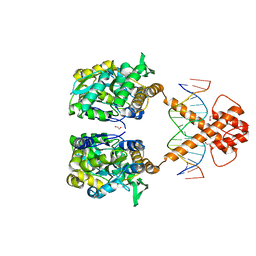 | | Crystal structure of MBP-fused BIL1/BZR1 (21-90) in complex with double-stranded DNA contaning CACACGTGTG | | Descriptor: | 1,2-ETHANEDIOL, DNA (5'-D(*TP*TP*CP*AP*CP*AP*CP*GP*TP*GP*TP*GP*AP*AP*A)-3'), Maltodextrin-binding protein,Protein BRASSINAZOLE-RESISTANT 1, ... | | Authors: | Nosaki, S, Tanokura, M, Miyakawa, T. | | Deposit date: | 2021-10-10 | | Release date: | 2022-12-07 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Brassinosteroid-induced gene repression requires specific and tight promoter binding of BIL1/BZR1 via DNA shape readout.
Nat.Plants, 8, 2022
|
|
7VN4
 
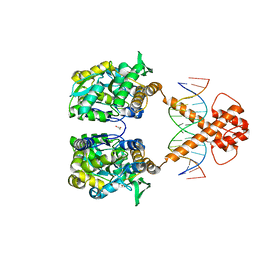 | | Crystal structure of MBP-fused BIL1/BZR1 (21-90) in complex with double-stranded DNA contaning TCCACGTGGA | | Descriptor: | 1,2-ETHANEDIOL, DNA (5'-D(*TP*TP*TP*CP*CP*AP*CP*GP*TP*GP*AP*AP*AP*AP*A)-3'), Maltodextrin-binding protein,Protein BRASSINAZOLE-RESISTANT 1, ... | | Authors: | Nosaki, S, Tanokura, M, Miyakawa, T. | | Deposit date: | 2021-10-10 | | Release date: | 2022-12-07 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Brassinosteroid-induced gene repression requires specific and tight promoter binding of BIL1/BZR1 via DNA shape readout.
Nat.Plants, 8, 2022
|
|
7VN5
 
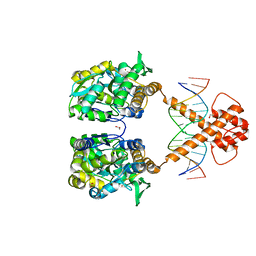 | | Crystal structure of MBP-fused BIL1/BZR1 (21-90) in complex with double-stranded DNA contaning TTCACGTGAA | | Descriptor: | 1,2-ETHANEDIOL, DNA (5'-D(*TP*TP*TP*TP*CP*AP*CP*GP*TP*GP*AP*AP*AP*AP*A)-3'), Maltodextrin-binding protein,Protein BRASSINAZOLE-RESISTANT 1, ... | | Authors: | Nosaki, S, Tanokura, M, Miyakawa, T. | | Deposit date: | 2021-10-10 | | Release date: | 2022-12-07 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Brassinosteroid-induced gene repression requires specific and tight promoter binding of BIL1/BZR1 via DNA shape readout.
Nat.Plants, 8, 2022
|
|
3VJN
 
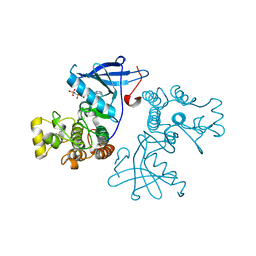 | | Crystal structure of the mutated EGFR kinase domain (G719S/T790M) in complex with AMPPNP. | | Descriptor: | Epidermal growth factor receptor, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER | | Authors: | Yoshikawa, S, Kukimoto-Niino, M, Shirouzu, M, Semba, K, Yamamoto, T, Yokoyama, S. | | Deposit date: | 2011-10-27 | | Release date: | 2012-03-07 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | Structural basis for the altered drug sensitivities of non-small cell lung cancer-associated mutants of human epidermal growth factor receptor.
Oncogene, 32, 2013
|
|
5XAX
 
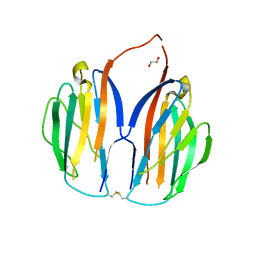 | |
5XAW
 
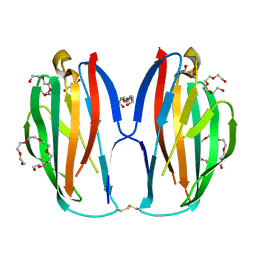 | | Parallel homodimer structures of voltage-gated sodium channel beta4 for cell-cell adhesion | | Descriptor: | 3,6,9,12,15,18-HEXAOXAICOSANE-1,20-DIOL, GLYCEROL, Sodium channel subunit beta-4, ... | | Authors: | Shimizu, H, Yokoyama, S. | | Deposit date: | 2017-03-15 | | Release date: | 2017-07-05 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.101 Å) | | Cite: | Parallel homodimer structures of the extracellular domains of the voltage-gated sodium channel beta 4 subunit explain its role in cell-cell adhesion
J. Biol. Chem., 292, 2017
|
|
2RR4
 
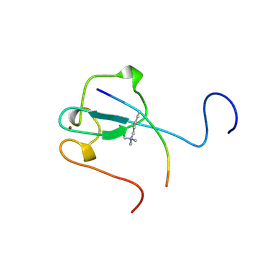 | | Complex structure of the zf-CW domain and the H3K4me3 peptide | | Descriptor: | Histone H3, ZINC ION, Zinc finger CW-type PWWP domain protein 1 | | Authors: | He, F, Muto, Y, Inoue, M, Kigawa, T, Shirouzu, M, Terada, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2010-03-24 | | Release date: | 2010-09-15 | | Last modified: | 2011-07-13 | | Method: | SOLUTION NMR | | Cite: | Structural insight into the zinc finger CW domain as a histone modification reader
Structure, 18, 2010
|
|
3VJO
 
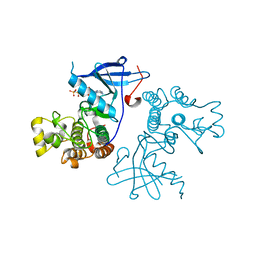 | | Crystal structure of the wild-type EGFR kinase domain in complex with AMPPNP. | | Descriptor: | Epidermal growth factor receptor, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER | | Authors: | Yoshikawa, S, Kukimoto-Niino, M, Shirouzu, M, Semba, K, Yamamoto, T, Yokoyama, S. | | Deposit date: | 2011-10-27 | | Release date: | 2012-03-07 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.64 Å) | | Cite: | Structural basis for the altered drug sensitivities of non-small cell lung cancer-associated mutants of human epidermal growth factor receptor.
Oncogene, 32, 2013
|
|
7VHC
 
 | | Crystal structure of the STX2a complexed with AR4A peptide | | Descriptor: | 3-PYRIDINIUM-1-YLPROPANE-1-SULFONATE, Shiga toxin 2 B subunit, inhibitor peptide, ... | | Authors: | Senda, M, Takahashi, M, Nishikawa, K, Senda, T. | | Deposit date: | 2021-09-22 | | Release date: | 2022-07-20 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | A unique peptide-based pharmacophore identifies an inhibitory compound against the A-subunit of Shiga toxin.
Sci Rep, 12, 2022
|
|
7VHE
 
 | | Crystal structure of the STX2a complexed with RRRA peptide | | Descriptor: | 3-PYRIDINIUM-1-YLPROPANE-1-SULFONATE, GLYCEROL, RRRA peptide, ... | | Authors: | Senda, M, Takahashi, M, Nishikawa, K, Senda, T. | | Deposit date: | 2021-09-22 | | Release date: | 2022-07-20 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | A unique peptide-based pharmacophore identifies an inhibitory compound against the A-subunit of Shiga toxin.
Sci Rep, 12, 2022
|
|
7VHF
 
 | | Crystal structure of the STX2a complexed with RRA peptide | | Descriptor: | 3-PYRIDINIUM-1-YLPROPANE-1-SULFONATE, GLYCEROL, RRA peptide, ... | | Authors: | Senda, M, Takahashi, M, Nishikawa, K, Senda, T. | | Deposit date: | 2021-09-22 | | Release date: | 2022-07-20 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | A unique peptide-based pharmacophore identifies an inhibitory compound against the A-subunit of Shiga toxin.
Sci Rep, 12, 2022
|
|
7VHD
 
 | | Crystal structure of the STX2a complexed with R4A peptide | | Descriptor: | 3-PYRIDINIUM-1-YLPROPANE-1-SULFONATE, ARG-ARG-ARG-ARG-ALA, Shiga toxin 2 B subunit, ... | | Authors: | Senda, M, Takahashi, M, Nishikawa, K, Senda, T. | | Deposit date: | 2021-09-22 | | Release date: | 2022-07-20 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | A unique peptide-based pharmacophore identifies an inhibitory compound against the A-subunit of Shiga toxin.
Sci Rep, 12, 2022
|
|
1WJ0
 
 | |
1WIJ
 
 | | Solution Structure of the DNA-Binding Domain of Ethylene-Insensitive3-Like3 | | Descriptor: | ETHYLENE-INSENSITIVE3-like 3 protein | | Authors: | Yamasaki, K, Inoue, M, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2004-05-28 | | Release date: | 2004-11-28 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the major DNA-binding domain of Arabidopsis thaliana ethylene-insensitive3-like3.
J.Mol.Biol., 348, 2005
|
|
7VOT
 
 | | The structure of dimeric photosynthetic RC-LH1 supercomplex in Class-2 | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, BACTERIOCHLOROPHYLL A, BACTERIOPHEOPHYTIN B, ... | | Authors: | Cao, P, Li, M, Liu, L.N. | | Deposit date: | 2021-10-14 | | Release date: | 2022-04-27 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structural basis for the assembly and quinone transport mechanisms of the dimeric photosynthetic RC-LH1 supercomplex.
Nat Commun, 13, 2022
|
|
7VOR
 
 | | The structure of dimeric photosynthetic RC-LH1 supercomplex in Class-1 | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, BACTERIOCHLOROPHYLL A, BACTERIOPHEOPHYTIN A, ... | | Authors: | Cao, P, Li, M, Liu, L.N. | | Deposit date: | 2021-10-14 | | Release date: | 2022-04-27 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (2.74 Å) | | Cite: | Structural basis for the assembly and quinone transport mechanisms of the dimeric photosynthetic RC-LH1 supercomplex.
Nat Commun, 13, 2022
|
|
7VB9
 
 | | Rba sphaeroides PufY-KO RC-LH1 dimer type-2 | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, BACTERIOCHLOROPHYLL A, BACTERIOPHEOPHYTIN A, ... | | Authors: | Bracun, L, Yamagata, A, Liu, L.N, Shirouzu, M. | | Deposit date: | 2021-08-30 | | Release date: | 2022-05-04 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.45 Å) | | Cite: | Structural basis for the assembly and quinone transport mechanisms of the dimeric photosynthetic RC-LH1 supercomplex.
Nat Commun, 13, 2022
|
|
7VOY
 
 | | Rba sphaeroides PufX-KO RC-LH1 | | Descriptor: | BACTERIOCHLOROPHYLL A, BACTERIOPHEOPHYTIN A, FE (II) ION, ... | | Authors: | Bracun, L, Yamagata, A, Liu, L.N, Shirouzu, M. | | Deposit date: | 2021-10-15 | | Release date: | 2022-05-04 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Structural basis for the assembly and quinone transport mechanisms of the dimeric photosynthetic RC-LH1 supercomplex.
Nat Commun, 13, 2022
|
|
7VA9
 
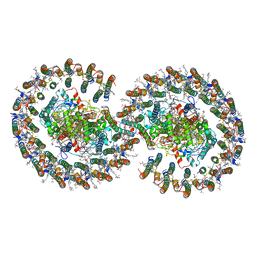 | | Rba sphaeroides PufY-KO RC-LH1 dimer type-1 | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, BACTERIOCHLOROPHYLL A, BACTERIOPHEOPHYTIN B, ... | | Authors: | Bracun, L, Yamagata, A, Liu, L.N, Shirouzu, M. | | Deposit date: | 2021-08-27 | | Release date: | 2022-05-04 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.08 Å) | | Cite: | Structural basis for the assembly and quinone transport mechanisms of the dimeric photosynthetic RC-LH1 supercomplex.
Nat Commun, 13, 2022
|
|
7VNM
 
 | | Rba sphaeroides PufY-KO RC-LH1 monomer | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, BACTERIOCHLOROPHYLL A, BACTERIOPHEOPHYTIN A, ... | | Authors: | Bracun, L, Yamagata, A, Liu, L.N, Shirouzu, M. | | Deposit date: | 2021-10-11 | | Release date: | 2022-05-04 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (2.86 Å) | | Cite: | Structural basis for the assembly and quinone transport mechanisms of the dimeric photosynthetic RC-LH1 supercomplex.
Nat Commun, 13, 2022
|
|
7VNY
 
 | | Rba sphaeroides WT RC-LH1 monomer | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, BACTERIOCHLOROPHYLL A, BACTERIOPHEOPHYTIN B, ... | | Authors: | Bracun, L, Yamagata, A, Liu, L.N, Shirouzu, M. | | Deposit date: | 2021-10-12 | | Release date: | 2022-06-01 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (2.79 Å) | | Cite: | Structural basis for the assembly and quinone transport mechanisms of the dimeric photosynthetic RC-LH1 supercomplex.
Nat Commun, 13, 2022
|
|
1WJ2
 
 | | Solution Structure of the C-terminal WRKY Domain of AtWRKY4 | | Descriptor: | Probable WRKY transcription factor 4, ZINC ION | | Authors: | Yamasaki, K, Inoue, M, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2004-05-28 | | Release date: | 2004-11-28 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of an Arabidopsis WRKY DNA binding domain.
Plant Cell, 17, 2005
|
|
1WID
 
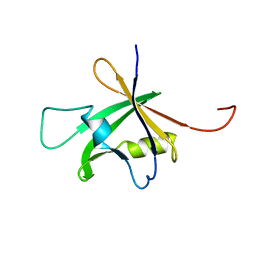 | | Solution Structure of the B3 DNA-Binding Domain of RAV1 | | Descriptor: | DNA-binding protein RAV1 | | Authors: | Yamasaki, K, Inoue, M, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2004-05-28 | | Release date: | 2004-11-28 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of the B3 DNA Binding Domain of the Arabidopsis Cold-Responsive Transcription Factor RAV1
Plant Cell, 16, 2004
|
|
1WVO
 
 | | Solution structure of RSGI RUH-029, an antifreeze protein like domain in human N-acetylneuraminic acid phosphate synthase gene. | | Descriptor: | Sialic acid synthase | | Authors: | Ito, Y, Hamada, T, Hayashi, F, Yokoyama, S, Hirota, H, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2004-12-22 | | Release date: | 2006-01-03 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the antifreeze-like domain of human sialic acid synthase
Protein Sci., 15, 2006
|
|
1WWQ
 
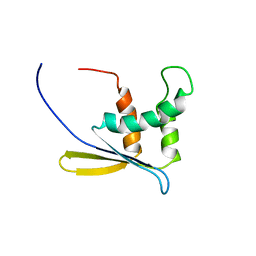 | | Solution Structure of Mouse ER | | Descriptor: | Enhancer of rudimentary homolog | | Authors: | Li, H, Koshiba, S, Inoue, M, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-01-12 | | Release date: | 2006-01-03 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the mouse enhancer of rudimentary protein reveals a novel fold
J.Biomol.Nmr, 32, 2005
|
|
