2YLI
 
 | |
2YL1
 
 | |
5DJE
 
 | | Crystal structure of the zuotin homology domain (ZHD) from yeast Zuo1 | | Descriptor: | 1,2-ETHANEDIOL, 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Shrestha, O.K, Bingman, C.A, Craig, E.A. | | Deposit date: | 2015-09-02 | | Release date: | 2016-09-28 | | Last modified: | 2019-12-25 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Dual interaction of the Hsp70 J-protein cochaperone Zuotin with the 40S and 60S ribosomal subunits.
Nat.Struct.Mol.Biol., 23, 2016
|
|
5KM4
 
 | |
5KLZ
 
 | |
5KM5
 
 | | Human Histidine Triad Nucleotide Binding Protein 2 (hHint2) triciribine 5'-monoposphate catalytic product complex | | Descriptor: | 5-methyl-1-(5-O-phosphono-beta-D-ribofuranosyl)-1,5-dihydro-1,4,5,6,8-pentaazaacenaphthylen-3-amine, CHLORIDE ION, Histidine triad nucleotide-binding protein 2, ... | | Authors: | Maize, K.M, Finzel, B.C. | | Deposit date: | 2016-06-26 | | Release date: | 2017-06-28 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | A Crystal Structure Based Guide to the Design of Human Histidine Triad Nucleotide Binding Protein 1 (hHint1) Activated ProTides.
Mol. Pharm., 14, 2017
|
|
5D54
 
 | | In meso X-ray crystallography structure of insulin at 100 K | | Descriptor: | 3,6,9,12,15,18,21,24-OCTAOXAHEXACOSAN-1-OL, Insulin A chain, Insulin B chain, ... | | Authors: | Huang, C.-Y, Olieric, V, Wang, M, Caffrey, M. | | Deposit date: | 2015-08-10 | | Release date: | 2016-01-13 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | In meso in situ serial X-ray crystallography of soluble and membrane proteins at cryogenic temperatures.
Acta Crystallogr D Struct Biol, 72, 2016
|
|
5D57
 
 | | In meso X-ray crystallography structure of diacylglycerol kinase, DgkA, at 100 K | | Descriptor: | (2S)-2,3-DIHYDROXYPROPYL(7Z)-PENTADEC-7-ENOATE, Diacylglycerol kinase | | Authors: | Huang, C.-Y, Howe, N, Olieric, V, Wang, M, Caffrey, M. | | Deposit date: | 2015-08-10 | | Release date: | 2016-01-13 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | In meso in situ serial X-ray crystallography of soluble and membrane proteins at cryogenic temperatures.
Acta Crystallogr D Struct Biol, 72, 2016
|
|
5D5F
 
 | | In meso in situ serial X-ray crystallography structure of lysozyme by bromine-SAD at 100 K | | Descriptor: | 3,6,9,12,15,18,21,24-OCTAOXAHEXACOSAN-1-OL, ACETIC ACID, BROMIDE ION, ... | | Authors: | Huang, C.-Y, Olieric, V, Diederichs, K, Wang, M, Caffrey, M. | | Deposit date: | 2015-08-10 | | Release date: | 2016-01-13 | | Last modified: | 2016-03-02 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | In meso in situ serial X-ray crystallography of soluble and membrane proteins at cryogenic temperatures.
Acta Crystallogr D Struct Biol, 72, 2016
|
|
5D53
 
 | | In meso in situ serial X-ray crystallography structure of insulin at 100 K | | Descriptor: | 3,6,9,12,15,18,21,24-OCTAOXAHEXACOSAN-1-OL, Insulin A chain, Insulin B chain, ... | | Authors: | Huang, C.-Y, Olieric, V, Diederichs, K, Wang, M, Caffrey, M. | | Deposit date: | 2015-08-10 | | Release date: | 2016-01-13 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | In meso in situ serial X-ray crystallography of soluble and membrane proteins at cryogenic temperatures.
Acta Crystallogr D Struct Biol, 72, 2016
|
|
5D5B
 
 | | In meso X-ray crystallography structure of the Beta2-adrenergic receptor at 100 K | | Descriptor: | (2S)-1-(9H-Carbazol-4-yloxy)-3-(isopropylamino)propan-2-ol, 1,4-BUTANEDIOL, ACETAMIDE, ... | | Authors: | Huang, C.-Y, Olieric, V, Liu, X, Kobilka, B, Wang, M, Caffrey, M. | | Deposit date: | 2015-08-10 | | Release date: | 2016-01-13 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.8 Å) | | Cite: | In meso in situ serial X-ray crystallography of soluble and membrane proteins at cryogenic temperatures.
Acta Crystallogr D Struct Biol, 72, 2016
|
|
2YL3
 
 | | CYTOCHROME C PRIME FROM ALCALIGENES XYLOSOXIDANS: CARBON MONOOXIDE BOUND L16G VARIANT AT 1.04 A RESOLUTION - RESTRAINT REFINED | | Descriptor: | CARBON MONOXIDE, CYTOCHROME C', HEME C, ... | | Authors: | Antonyuk, S.V, Rustage, N, Eady, R.R, Hasnain, S.S. | | Deposit date: | 2011-05-31 | | Release date: | 2011-10-05 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.04 Å) | | Cite: | Carbon Monoxide Poisoning is Prevented by the Energy Costs of Conformational Changes in Gas- Binding Haemproteins.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
5KM9
 
 | |
2K1H
 
 | | Solution NMR structure of SeR13 from Staphylococcus epidermidis. Northeast Structural Genomics Consortium target SeR13 | | Descriptor: | Uncharacterized protein SeR13 | | Authors: | Lee, H, Wylie, G, Bansal, S, Wang, X, Shastry, R, Jiang, M, Cunningham, K, Ma, L, Xiao, R, Liu, J, Baran, M.C, Swapna, G, Acton, T.B, Rost, B, Montelione, G.T, Prestegard, J.H, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2008-03-05 | | Release date: | 2008-03-18 | | Last modified: | 2024-05-08 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of SeR13.
To be Published
|
|
5KM3
 
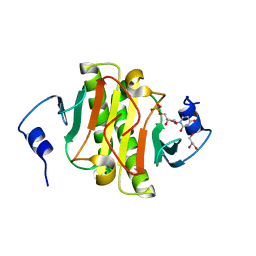 | |
5KM1
 
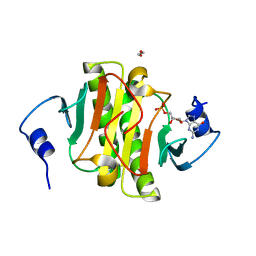 | |
5KMA
 
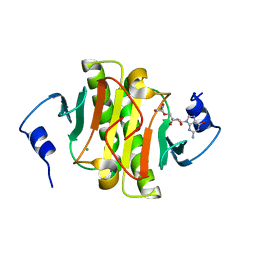 | | Human Histidine Triad Nucleotide Binding Protein 1 (hHint1) H112N mutant nucleoside D-Trp phosphoramidate substrate complex | | Descriptor: | CHLORIDE ION, Histidine triad nucleotide-binding protein 1, [(2~{R},3~{S},4~{R},5~{R})-5-(2-azanyl-6-oxidanylidene-3~{H}-purin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methoxy-~{N}-ethyl-phosphonamidic acid | | Authors: | Maize, K.M, Finzel, B.C. | | Deposit date: | 2016-06-26 | | Release date: | 2017-06-28 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | A Crystal Structure Based Guide to the Design of Human Histidine Triad Nucleotide Binding Protein 1 (hHint1) Activated ProTides.
Mol. Pharm., 14, 2017
|
|
5D58
 
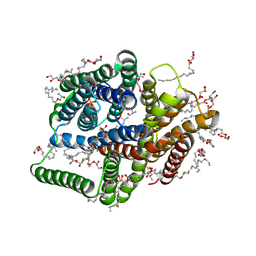 | | In meso in situ serial X-ray crystallography structure of the PepTSt-Ala-Phe complex at 100 K | | Descriptor: | (2S)-2,3-DIHYDROXYPROPYL(7Z)-PENTADEC-7-ENOATE, 3,6,9,12,15,18,21,24-OCTAOXAHEXACOSAN-1-OL, ALANINE, ... | | Authors: | Huang, C.-Y, Olieric, V, Diederichs, K, Wang, M, Caffrey, M. | | Deposit date: | 2015-08-10 | | Release date: | 2016-01-13 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | In meso in situ serial X-ray crystallography of soluble and membrane proteins at cryogenic temperatures.
Acta Crystallogr D Struct Biol, 72, 2016
|
|
5D5D
 
 | | In meso in situ serial X-ray crystallography structure of AlgE at 100 K | | Descriptor: | (2S)-2,3-DIHYDROXYPROPYL(7Z)-PENTADEC-7-ENOATE, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 3,6,9,12,15,18,21,24-OCTAOXAHEXACOSAN-1-OL, ... | | Authors: | Ma, P, Huang, C.-Y, Olieric, V, Diederichs, K, Wang, M, Caffrey, M. | | Deposit date: | 2015-08-10 | | Release date: | 2016-01-13 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | In meso in situ serial X-ray crystallography of soluble and membrane proteins at cryogenic temperatures.
Acta Crystallogr D Struct Biol, 72, 2016
|
|
2YLD
 
 | | RECOMBINANT NATIVE CYTOCHROME C PRIME FROM ALCALIGENES XYLOSOXIDANS: CARBON MONOOXIDE BOUND AT 1.25 A | | Descriptor: | ASCORBIC ACID, CARBON MONOXIDE, CYTOCHROME C', ... | | Authors: | Antonyuk, S.V, Rustage, N, Eady, R.R, Hasnain, S.S. | | Deposit date: | 2011-06-02 | | Release date: | 2011-10-05 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Carbon Monoxide Poisoning is Prevented by the Energy Costs of Conformational Changes in Gas- Binding Haemproteins.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
5D59
 
 | | In meso X-ray crystallography structure of the PepTSt-Ala-Phe complex at 100 K | | Descriptor: | (2S)-2,3-DIHYDROXYPROPYL(7Z)-PENTADEC-7-ENOATE, 3,6,9,12,15,18,21,24-OCTAOXAHEXACOSAN-1-OL, ALANINE, ... | | Authors: | Huang, C.-Y, Olieric, V, Wang, M, Caffrey, M. | | Deposit date: | 2015-08-10 | | Release date: | 2016-01-13 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | In meso in situ serial X-ray crystallography of soluble and membrane proteins at cryogenic temperatures.
Acta Crystallogr D Struct Biol, 72, 2016
|
|
5D5C
 
 | | In meso in situ serial X-ray crystallography structure of lysozyme at 100 K | | Descriptor: | 3,6,9,12,15,18,21,24-OCTAOXAHEXACOSAN-1-OL, ACETIC ACID, BROMIDE ION, ... | | Authors: | Huang, C.-Y, Olieric, V, Diederichs, K, Wang, M, Caffrey, M. | | Deposit date: | 2015-08-10 | | Release date: | 2016-01-13 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | In meso in situ serial X-ray crystallography of soluble and membrane proteins at cryogenic temperatures.
Acta Crystallogr D Struct Biol, 72, 2016
|
|
5KMB
 
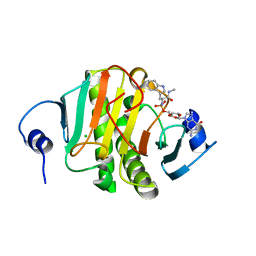 | | Human Histidine Triad Nucleotide Binding Protein 1 (hHint1) H112N mutant nucleoside L-Trp phosphoramidate substrate complex | | Descriptor: | CHLORIDE ION, Histidine triad nucleotide-binding protein 1, [(2~{R},3~{S},4~{R},5~{R})-5-(2-azanyl-6-oxidanylidene-1~{H}-purin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methoxy-~{N}-[(2~ {S})-3-(1~{H}-indol-3-yl)-1-(methylamino)-1-oxidanylidene-propan-2-yl]phosphonamidic acid | | Authors: | Maize, K.M, Finzel, B.C. | | Deposit date: | 2016-06-26 | | Release date: | 2017-06-28 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | A Crystal Structure Based Guide to the Design of Human Histidine Triad Nucleotide Binding Protein 1 (hHint1) Activated ProTides.
Mol. Pharm., 14, 2017
|
|
5KM6
 
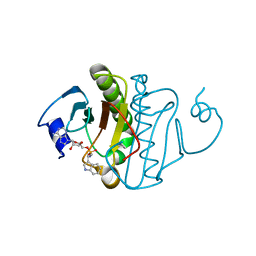 | | Human Histidine Triad Nucleotide Binding Protein 1 (hHint1) H112N mutant Ara-A nucleoside phosphoramidate substrate complex | | Descriptor: | Histidine triad nucleotide-binding protein 1, [(2~{R},3~{S},4~{S},5~{R})-5-(6-aminopurin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methoxy-~{N}-[2-(1~{H}-indol-3-yl)ethyl]phosphonamidic acid | | Authors: | Maize, K.M, Finzel, B.C. | | Deposit date: | 2016-06-26 | | Release date: | 2017-06-28 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | A Crystal Structure Based Guide to the Design of Human Histidine Triad Nucleotide Binding Protein 1 (hHint1) Activated ProTides.
Mol. Pharm., 14, 2017
|
|
3O0J
 
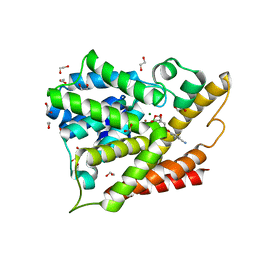 | | PDE4B In complex with ligand an2898 | | Descriptor: | 1,2-ETHANEDIOL, 4-[(1-hydroxy-1,3-dihydro-2,1-benzoxaborol-5-yl)oxy]benzene-1,2-dicarbonitrile, MAGNESIUM ION, ... | | Authors: | Alley, M.R.K, Zhou, Y. | | Deposit date: | 2010-07-19 | | Release date: | 2011-08-10 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Boron-based phosphodiesterase inhibitors show novel binding of boron to PDE4 bimetal center.
Febs Lett., 586, 2012
|
|
