8JI6
 
 | | Crystal structure of AetD in complex with L-tryptophan | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, AetD, FE (II) ION, ... | | Authors: | Li, H, Dai, L, Zheng, H.B, Chen, C.-C, Guo, R.-T. | | Deposit date: | 2023-05-26 | | Release date: | 2023-11-15 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The structural and functional investigation into an unusual nitrile synthase.
Nat Commun, 14, 2023
|
|
8JI2
 
 | | Crystal structure of AetD in complex with 5,7-dibromo-L-tryptophan | | Descriptor: | (2S)-2-azanyl-3-[5,7-bis(bromanyl)-1H-indol-3-yl]propanoic acid, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, AetD, ... | | Authors: | Li, H, Dai, L, Zheng, H.B, Chen, C.-C, Guo, R.-T. | | Deposit date: | 2023-05-26 | | Release date: | 2023-11-15 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | The structural and functional investigation into an unusual nitrile synthase.
Nat Commun, 14, 2023
|
|
8JI7
 
 | | Crystal structure of AetD in complex with L-tryptophan and two Fe2+ | | Descriptor: | AetD, FE (II) ION, NICKEL (II) ION, ... | | Authors: | Li, H, Dai, L, Zheng, H.B, Chen, C.-C, Guo, R.-T. | | Deposit date: | 2023-05-26 | | Release date: | 2023-11-15 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | The structural and functional investigation into an unusual nitrile synthase.
Nat Commun, 14, 2023
|
|
8J45
 
 | | Crystal structure of a Pichia pastoris-expressed IsPETase variant | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Poly(ethylene terephthalate) hydrolase | | Authors: | Li, X, He, H.L, Long, X, Niu, D, Huang, J.-W, Chen, C.-C, Guo, R.-T. | | Deposit date: | 2023-04-19 | | Release date: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Complete decomposition of poly(ethylene terephthalate) by crude PET hydrolytic enzyme produced in Pichia pastoris
Chem Eng J, 2023
|
|
6KQX
 
 | | Crystal structure of Yijc from B. subtilis in complex with UDP | | Descriptor: | URIDINE-5'-DIPHOSPHATE, Uncharacterized UDP-glucosyltransferase YjiC | | Authors: | Hu, Y.M, Dai, L.H, Huang, J.W, Liu, W.D, Chen, C.C, Guo, R.T. | | Deposit date: | 2019-08-20 | | Release date: | 2020-09-30 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.44 Å) | | Cite: | Structural dissection of unnatural ginsenoside-biosynthetic UDP-glycosyltransferase Bs-YjiC from Bacillus subtilis for substrate promiscuity.
Biochem.Biophys.Res.Commun., 534, 2021
|
|
6LAA
 
 | | Crystal structure of full-length CYP116B46 from Tepidiphilus thermophilus | | Descriptor: | 1,2-ETHANEDIOL, CARBONATE ION, Cytochrome P450, ... | | Authors: | Zhang, L.L, Ko, T.P, Huang, J.W, Liu, W.D, Chen, C.C, Guo, R.T. | | Deposit date: | 2019-11-12 | | Release date: | 2020-06-10 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.13 Å) | | Cite: | Structural insight into the electron transfer pathway of a self-sufficient P450 monooxygenase.
Nat Commun, 11, 2020
|
|
6KQW
 
 | | Crystal structure of Yijc from B. subtilis | | Descriptor: | CITRIC ACID, Uncharacterized UDP-glucosyltransferase YjiC | | Authors: | Hu, Y.M, Dai, L.H, Huang, J.W, Liu, W.D, Chen, C.C, Guo, R.T. | | Deposit date: | 2019-08-20 | | Release date: | 2020-09-30 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | Structural dissection of unnatural ginsenoside-biosynthetic UDP-glycosyltransferase Bs-YjiC from Bacillus subtilis for substrate promiscuity.
Biochem.Biophys.Res.Commun., 534, 2021
|
|
6LDL
 
 | | Crystal structure of CYP116B46-N(20-445) from Tepidiphilus thermophilus in complex with HEME | | Descriptor: | BICINE, Cytochrome P450, GLYCEROL, ... | | Authors: | Zhang, L.L, Xie, Z.Z, Huang, J.W, Liu, W.D, Chen, C.C, Guo, R.T. | | Deposit date: | 2019-11-21 | | Release date: | 2020-10-28 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.38 Å) | | Cite: | Structural insight into the electron transfer pathway of a self-sufficient P450 monooxygenase.
Nat Commun, 11, 2020
|
|
4DJW
 
 | |
4DJU
 
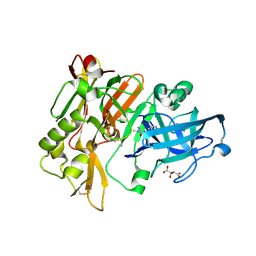 | | Structure of BACE Bound to 2-imino-3-methyl-5,5-diphenylimidazolidin-4-one | | Descriptor: | (2E)-2-imino-3-methyl-5,5-diphenylimidazolidin-4-one, Beta-secretase 1, L(+)-TARTARIC ACID | | Authors: | Strickland, C, Cumming, J. | | Deposit date: | 2012-02-02 | | Release date: | 2012-03-21 | | Last modified: | 2012-04-04 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure based design of iminohydantoin BACE1 inhibitors: Identification of an orally available, centrally active BACE1 inhibitor.
Bioorg.Med.Chem.Lett., 22, 2012
|
|
4DJX
 
 | |
3L59
 
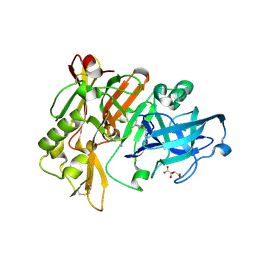 | | Structure of BACE Bound to SCH710413 | | Descriptor: | (2Z)-3-(3-chlorobenzyl)-2-imino-5,5-dimethylimidazolidin-4-one, Beta-secretase 1, D(-)-TARTARIC ACID | | Authors: | Strickland, C, Zhu, Z. | | Deposit date: | 2009-12-21 | | Release date: | 2010-02-16 | | Last modified: | 2018-01-24 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Discovery of Cyclic Acylguanidines as Highly Potent and Selective beta-Site Amyloid Cleaving Enzyme (BACE) Inhibitors: Part I-Inhibitor Design and Validation
J.Med.Chem., 53, 2010
|
|
3MP1
 
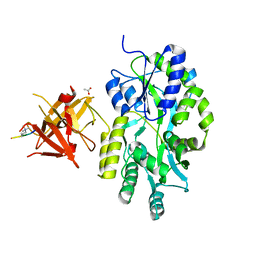 | | Complex structure of Sgf29 and trimethylated H3K4 | | Descriptor: | ACETATE ION, H3K4me3 peptide, Maltose-binding periplasmic protein,LINKER,SAGA-associated factor 29, ... | | Authors: | Li, J, Ruan, J, Wu, M, Xue, X, Zang, J. | | Deposit date: | 2010-04-24 | | Release date: | 2011-05-04 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Sgf29 binds histone H3K4me2/3 and is required for SAGA complex recruitment and histone H3 acetylation
Embo J., 30, 2011
|
|
3MP8
 
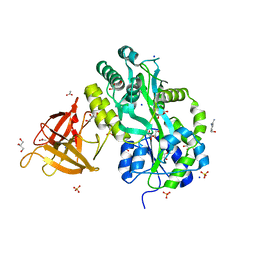 | | Crystal structure of Sgf29 tudor domain | | Descriptor: | 4-(HYDROXYMETHYL)BENZAMIDINE, ACETIC ACID, GLYCEROL, ... | | Authors: | Li, J, Wu, M, Ruan, J, Zang, J. | | Deposit date: | 2010-04-26 | | Release date: | 2011-05-04 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Sgf29 binds histone H3K4me2/3 and is required for SAGA complex recruitment and histone H3 acetylation
Embo J., 30, 2011
|
|
3MP6
 
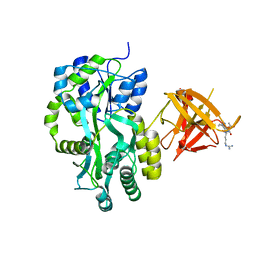 | | Complex Structure of Sgf29 and dimethylated H3K4 | | Descriptor: | H3K4me2 peptide, Maltose-binding periplasmic protein,LINKER,SAGA-associated factor 29, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Li, J, Wu, M, Ruan, J, Zang, J. | | Deposit date: | 2010-04-25 | | Release date: | 2011-05-04 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Sgf29 binds histone H3K4me2/3 and is required for SAGA complex recruitment and histone H3 acetylation
Embo J., 30, 2011
|
|
7WYI
 
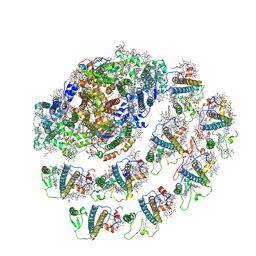 | | Native Photosystem I of Chlamydomonas reinhardtii | | Descriptor: | CHLOROPHYLL A, CHLOROPHYLL A ISOMER, CHLOROPHYLL B, ... | | Authors: | Kurisu, G, Gerle, C, Mitsuoka, K, Kawamoto, A, Tanaka, H. | | Deposit date: | 2022-02-16 | | Release date: | 2023-02-22 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Three structures of PSI-LHCI from Chlamydomonas reinhardtii suggest a resting state re-activated by ferredoxin.
Biochim Biophys Acta Bioenerg, 1864, 2023
|
|
7WZN
 
 | | PSI-LHCI from Chlamydomonas reinhardtii with bound ferredoxin | | Descriptor: | CHLOROPHYLL A, CHLOROPHYLL A ISOMER, CHLOROPHYLL B, ... | | Authors: | Kurisu, G, Gerle, C, Mitsuoka, K, Kawamoto, A, Tanaka, H. | | Deposit date: | 2022-02-18 | | Release date: | 2023-02-22 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (4.9 Å) | | Cite: | Three structures of PSI-LHCI from Chlamydomonas reinhardtii suggest a resting state re-activated by ferredoxin.
Biochim Biophys Acta Bioenerg, 1864, 2023
|
|
5ENH
 
 | | Crystal structure of the second bromodomain of Pleckstrin homology domain interacting protein (PHIP) in complex with compound-12 N11528 (SGC - Diamond I04-1 fragment screening) | | Descriptor: | PH-interacting protein, ~{N}-[(2,6-dimethoxyphenyl)methyl]ethanamide | | Authors: | Krojer, T, Talon, R, Collins, P, Bradley, A, Cox, O, Szykowska, A, Burgess-Brown, N, Brennan, P, Bountra, C, Arrowsmith, C.H, Edwards, A, von Delft, F, Structural Genomics Consortium (SGC) | | Deposit date: | 2015-11-09 | | Release date: | 2016-04-27 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | A poised fragment library enables rapid synthetic expansion yielding the first reported inhibitors of PHIP(2), an atypical bromodomain.
Chem Sci, 7, 2016
|
|
5ENB
 
 | | Crystal structure of the second bromodomain of Pleckstrin homology domain interacting protein (PHIP) in complex with o-Tolylthiourea (SGC - Diamond I04-1 fragment screening) | | Descriptor: | 1,2-ETHANEDIOL, 1-(2-methylphenyl)thiourea, PH-interacting protein | | Authors: | Krojer, T, Talon, R, Collins, P, Bradley, A, Cox, O, Szykowska, A, Burgess-Brown, N, Brennan, P, Bountra, C, Arrowsmith, C.H, Edwards, A, von Delft, F, Structural Genomics Consortium (SGC) | | Deposit date: | 2015-11-09 | | Release date: | 2016-04-27 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | A poised fragment library enables rapid synthetic expansion yielding the first reported inhibitors of PHIP(2), an atypical bromodomain.
Chem Sci, 7, 2016
|
|
5ENI
 
 | | Crystal structure of the second bromodomain of Pleckstrin homology domain interacting protein (PHIP) in complex with compound-13 N11537 (SGC - Diamond I04-1 fragment screening) | | Descriptor: | PH-interacting protein, ~{N}-[[2,6-bis(chloranyl)phenyl]methyl]-2-oxidanyl-ethanamide | | Authors: | Krojer, T, Talon, R, Collins, P, Bradley, A, Cox, O, Szykowska, A, Burgess-Brown, N, Brennan, P, Bountra, C, Arrowsmith, C.H, Edwards, A, von Delft, F, Structural Genomics Consortium (SGC) | | Deposit date: | 2015-11-09 | | Release date: | 2016-04-27 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | A poised fragment library enables rapid synthetic expansion yielding the first reported inhibitors of PHIP(2), an atypical bromodomain.
Chem Sci, 7, 2016
|
|
5ENJ
 
 | | Crystal structure of the second bromodomain of Pleckstrin homology domain interacting protein (PHIP) in complex with compound-14 N11530 (SGC - Diamond I04-1 fragment screening) | | Descriptor: | MAGNESIUM ION, PH-interacting protein, ~{N}-[[2,6-bis(chloranyl)phenyl]methyl]-~{N}-methyl-ethanamide | | Authors: | Krojer, T, Talon, R, Collins, P, Bradley, A, Cox, O, Szykowska, A, Burgess-Brown, N, Brennan, P, Bountra, C, Arrowsmith, C.H, Edwards, A, von Delft, F, Structural Genomics Consortium (SGC) | | Deposit date: | 2015-11-09 | | Release date: | 2016-04-27 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | A poised fragment library enables rapid synthetic expansion yielding the first reported inhibitors of PHIP(2), an atypical bromodomain.
Chem Sci, 7, 2016
|
|
6XKC
 
 | | Crystal structure of E3 ligase | | Descriptor: | Protein fem-1 homolog C | | Authors: | Yan, X, Dong, A, Bountra, C, Edwards, A.M, Arrowsmith, C.H, Min, J.R, Dong, C, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-06-26 | | Release date: | 2020-10-14 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Molecular basis for ubiquitin ligase CRL2 FEM1C -mediated recognition of C-degron.
Nat.Chem.Biol., 17, 2021
|
|
6KC0
 
 | | fused To-MtbCsm1 with 2ATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, CRISPR system single-strand-specific deoxyribonuclease Cas10/Csm1 (subtype III-A),CRISPR system single-strand-specific deoxyribonuclease Cas10/Csm1 (subtype III-A), MAGNESIUM ION | | Authors: | Li, T, Huo, Y, Jiang, T. | | Deposit date: | 2019-06-26 | | Release date: | 2020-07-01 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.295 Å) | | Cite: | Mycobacterium tuberculosis CRISPR/Cas system Csm1 holds clues to the evolutionary relationship between DNA polymerase and cyclase activity.
Int.J.Biol.Macromol., 170, 2020
|
|
6PI7
 
 | |
6WAT
 
 | | complex structure of PHF1 | | Descriptor: | Histone H3.1t peptide, PHD finger protein 1, UNKNOWN ATOM OR ION | | Authors: | Dong, C, Bountra, C, Edwards, A.M, Arrowsmith, C.H, Min, J.R, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-03-26 | | Release date: | 2020-08-26 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural basis for histone variant H3tK27me3 recognition by PHF1 and PHF19.
Elife, 9, 2020
|
|
