8IW6
 
 | | crystal structure of SulE mutant | | Descriptor: | Alpha/beta fold hydrolase, GLYCEROL, L(+)-TARTARIC ACID, ... | | Authors: | Liu, B, He, J, Ran, T, Wang, W. | | Deposit date: | 2023-03-29 | | Release date: | 2023-08-02 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.44 Å) | | Cite: | Crystal structures of herbicide-detoxifying esterase reveal a lid loop affecting substrate binding and activity.
Nat Commun, 14, 2023
|
|
8IVE
 
 | | crystal structure of SulE mutant | | Descriptor: | 2-[[[[(4-CHLORO-6-METHOXY-2-PYRIMIDINYL)AMINO]CARBONYL]AMINO]SULFONYL]BENZOIC ACID ETHYL ESTER, Alpha/beta fold hydrolase, GLYCEROL | | Authors: | Liu, B, He, J, Ran, T, Wang, W. | | Deposit date: | 2023-03-27 | | Release date: | 2023-08-02 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.44 Å) | | Cite: | Crystal structures of herbicide-detoxifying esterase reveal a lid loop affecting substrate binding and activity.
Nat Commun, 14, 2023
|
|
8IVN
 
 | | crystal structure of SulE mutant | | Descriptor: | Alpha/beta fold hydrolase, GLYCEROL, L(+)-TARTARIC ACID, ... | | Authors: | Liu, B, He, J, Ran, T, Wang, W. | | Deposit date: | 2023-03-28 | | Release date: | 2023-08-02 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structures of herbicide-detoxifying esterase reveal a lid loop affecting substrate binding and activity.
Nat Commun, 14, 2023
|
|
8IVT
 
 | | crystal structure of SulE mutant | | Descriptor: | Alpha/beta fold hydrolase, GLYCEROL, L(+)-TARTARIC ACID, ... | | Authors: | Liu, B, He, J, Ran, T, Wang, W. | | Deposit date: | 2023-03-28 | | Release date: | 2023-08-02 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | Crystal structures of herbicide-detoxifying esterase reveal a lid loop affecting substrate binding and activity.
Nat Commun, 14, 2023
|
|
8IW3
 
 | | crystal structure of SulE mutant | | Descriptor: | 2-[[[[(4-CHLORO-6-METHOXY-2-PYRIMIDINYL)AMINO]CARBONYL]AMINO]SULFONYL]BENZOIC ACID ETHYL ESTER, Alpha/beta fold hydrolase, GLYCEROL, ... | | Authors: | Liu, B, He, J, Ran, T, Wang, W. | | Deposit date: | 2023-03-29 | | Release date: | 2023-08-02 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | Crystal structures of herbicide-detoxifying esterase reveal a lid loop affecting substrate binding and activity.
Nat Commun, 14, 2023
|
|
8J7G
 
 | | crystal structure of SulE mutant | | Descriptor: | Alpha/beta fold hydrolase, GLYCEROL, L(+)-TARTARIC ACID, ... | | Authors: | Liu, B, He, J, Ran, T, Wang, W. | | Deposit date: | 2023-04-27 | | Release date: | 2023-08-02 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Crystal structures of herbicide-detoxifying esterase reveal a lid loop affecting substrate binding and activity.
Nat Commun, 14, 2023
|
|
8J7K
 
 | | crystal structure of SulE mutant | | Descriptor: | Alpha/beta fold hydrolase, GLYCEROL, L(+)-TARTARIC ACID, ... | | Authors: | Liu, B, He, J, Ran, T, Wang, W. | | Deposit date: | 2023-04-27 | | Release date: | 2023-08-02 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.36 Å) | | Cite: | Crystal structures of herbicide-detoxifying esterase reveal a lid loop affecting substrate binding and activity.
Nat Commun, 14, 2023
|
|
7VOP
 
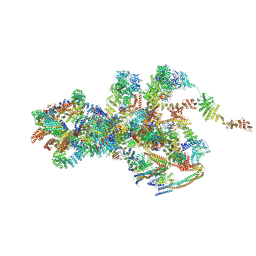 | | Cryo-EM structure of Xenopus laevis nuclear pore complex cytoplasmic ring subunit | | Descriptor: | GATOR complex protein SEC13, IL4I1 protein, MGC154553 protein, ... | | Authors: | Tai, L, Zhu, Y, Sun, F. | | Deposit date: | 2021-10-14 | | Release date: | 2022-02-02 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (8.7 Å) | | Cite: | 8 angstrom structure of the outer rings of the Xenopus laevis nuclear pore complex obtained by cryo-EM and AI.
Protein Cell, 13, 2022
|
|
7VCI
 
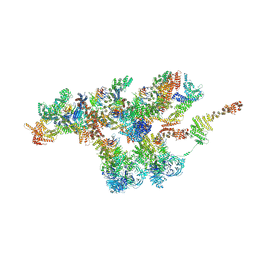 | | Structure of Xenopus laevis NPC nuclear ring asymmetric unit | | Descriptor: | GATOR complex protein SEC13, MGC154553 protein, MGC83295 protein, ... | | Authors: | Tai, L, Zhu, Y, Sun, F. | | Deposit date: | 2021-09-03 | | Release date: | 2022-02-02 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (8.1 Å) | | Cite: | 8 angstrom structure of the outer rings of the Xenopus laevis nuclear pore complex obtained by cryo-EM and AI.
Protein Cell, 13, 2022
|
|
7E7D
 
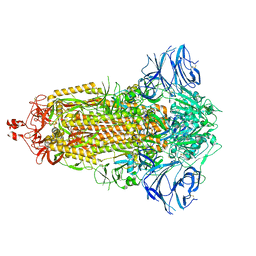 | |
7E7B
 
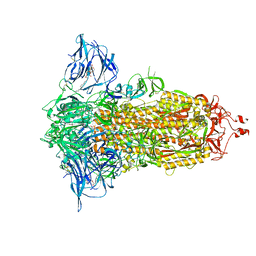 | | Cryo-EM structure of the SARS-CoV-2 furin site mutant S-Trimer from a subunit vaccine candidate | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-hydroxyethyl 2-deoxy-3,5-bis-O-(2-hydroxyethyl)-6-O-(2-{[(9E)-octadec-9-enoyl]oxy}ethyl)-alpha-L-xylo-hexofuranoside, ... | | Authors: | Zheng, S, Ma, J. | | Deposit date: | 2021-02-25 | | Release date: | 2021-03-24 | | Last modified: | 2022-12-07 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Cryo-EM structure of S-Trimer, a subunit vaccine candidate for COVID-19.
J.Virol., 95, 2021
|
|
7WWH
 
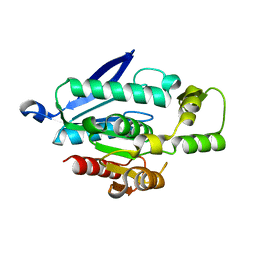 | |
8H0M
 
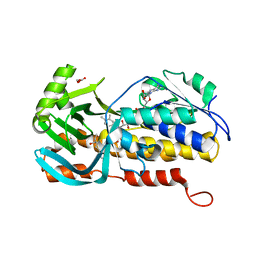 | | Crystal structure of VioD | | Descriptor: | (2S)-2-ethylhexan-1-ol, FLAVIN-ADENINE DINUCLEOTIDE, FORMIC ACID, ... | | Authors: | Xu, M, Ran, T, Wang, W. | | Deposit date: | 2022-09-29 | | Release date: | 2023-04-12 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.702 Å) | | Cite: | Structural basis for substrate binding and catalytic mechanism of the key enzyme VioD in the violacein synthesis pathway.
Proteins, 91, 2023
|
|
8J1V
 
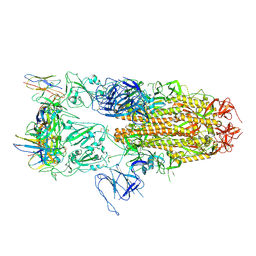 | |
8J1T
 
 | |
7EN0
 
 | | Structure and Activity of SLAC1 Channels for Stomatal Signaling in Leaves | | Descriptor: | DIUNDECYL PHOSPHATIDYL CHOLINE, SLow Anion Channel 1, SPHINGOSINE | | Authors: | Deng, Y, Kashtoh, H, Wang, Q, Zhen, G, Li, Q, Tang, L, Gao, H, Zhang, C, Qin, L, Su, M, Li, F, Huang, X, Wang, Y, Xie, Q, Clarke, O.B, Hendrickson, W.A, Chen, Y. | | Deposit date: | 2021-04-15 | | Release date: | 2021-05-19 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (2.97 Å) | | Cite: | Structure and activity of SLAC1 channels for stomatal signaling in leaves.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7DHT
 
 | |
