7BV6
 
 | | Crystal structure of the autophagic STX17/SNAP29/VAMP8 SNARE complex | | Descriptor: | Synaptosomal-associated protein 29, Syntaxin-17, Vesicle-associated membrane protein 8 | | Authors: | Li, Y, Pan, L.F. | | Deposit date: | 2020-04-09 | | Release date: | 2020-09-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.05 Å) | | Cite: | Decoding three distinct states of the Syntaxin17 SNARE motif in mediating autophagosome-lysosome fusion.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
7BV4
 
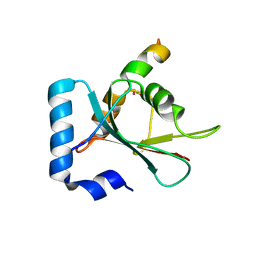 | | Crystal structure of STX17 LIR region in complex with GABARAP | | Descriptor: | DI(HYDROXYETHYL)ETHER, GLYCEROL, Gamma-aminobutyric acid receptor-associated protein, ... | | Authors: | Li, Y, Pan, L.F. | | Deposit date: | 2020-04-09 | | Release date: | 2020-09-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Decoding three distinct states of the Syntaxin17 SNARE motif in mediating autophagosome-lysosome fusion.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
7VZN
 
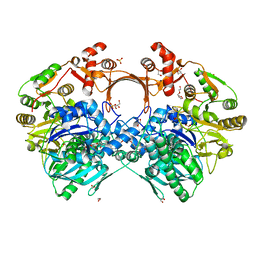 | | The structure of GdmN in complex with carbamoyl adenylate intermediate and 20-O-methyl-19-chloroproansamitocin | | Descriptor: | (5~{S},6~{E},8~{S},9~{S},12~{R},15~{E})-21-chloranyl-12,20-dimethoxy-6,8,16-trimethyl-5,9-bis(oxidanyl)-2-azabicyclo[16.3.1]docosa-1(21),6,15,18(22),19-pentaene-3,11-dione, 1,2-ETHANEDIOL, 5'-O-[(S)-(carbamoyloxy)(hydroxy)phosphoryl]adenosine, ... | | Authors: | Wei, J, Zheng, J, Zhou, J, Kang, Q, Bai, L. | | Deposit date: | 2021-11-16 | | Release date: | 2022-11-16 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Endowing homodimeric carbamoyltransferase GdmN with iterative functions through structural characterization and mechanistic studies.
Nat Commun, 13, 2022
|
|
7VZQ
 
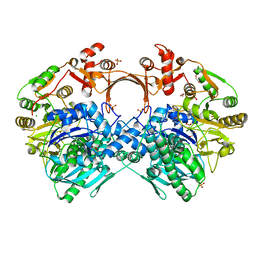 | | The structure of GdmN V24Y/G157A/R158A/G188R mutant in complex with carbamoyl adenylate intermediate | | Descriptor: | 1,2-ETHANEDIOL, 5'-O-[(S)-(carbamoyloxy)(hydroxy)phosphoryl]adenosine, CHLORIDE ION, ... | | Authors: | Wei, J, Zheng, J, Zhou, J, Kang, Q, Bai, L. | | Deposit date: | 2021-11-16 | | Release date: | 2022-11-16 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Endowing homodimeric carbamoyltransferase GdmN with iterative functions through structural characterization and mechanistic studies.
Nat Commun, 13, 2022
|
|
7VZU
 
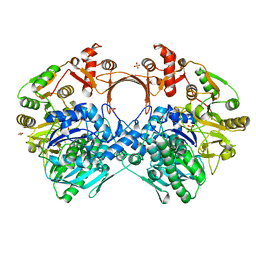 | | The structure of GdmN Y82F mutant | | Descriptor: | 1,2-ETHANEDIOL, FE (III) ION, GdmN, ... | | Authors: | Wei, J, Zheng, J, Zhou, J, Kang, Q, Bai, L. | | Deposit date: | 2021-11-16 | | Release date: | 2022-11-16 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Endowing homodimeric carbamoyltransferase GdmN with iterative functions through structural characterization and mechanistic studies.
Nat Commun, 13, 2022
|
|
7VX0
 
 | | The structure of GdmN complex with ATP | | Descriptor: | 1,2-ETHANEDIOL, ADENOSINE-5'-TRIPHOSPHATE, FE (III) ION, ... | | Authors: | Wei, J, Zheng, J, Zhou, J, Kang, Q, Bai, L. | | Deposit date: | 2021-11-12 | | Release date: | 2022-11-16 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Endowing homodimeric carbamoyltransferase GdmN with iterative functions through structural characterization and mechanistic studies.
Nat Commun, 13, 2022
|
|
7VYP
 
 | | The structure of GdmN complex with the natural tetrahedral intermediate, carbamoylated derivative, and AMP | | Descriptor: | 1,2-ETHANEDIOL, ADENOSINE MONOPHOSPHATE, FE (III) ION, ... | | Authors: | Wei, J, Zheng, J, Zhou, J, Kang, Q, Bai, L. | | Deposit date: | 2021-11-15 | | Release date: | 2022-11-16 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.88 Å) | | Cite: | Endowing homodimeric carbamoyltransferase GdmN with iterative functions through structural characterization and mechanistic studies.
Nat Commun, 13, 2022
|
|
7VZY
 
 | | The structure of GdmN complex with AMP and 20-O-methyl-19-chloroproansamitocin | | Descriptor: | (5~{S},6~{E},8~{S},9~{S},12~{R},15~{E})-21-chloranyl-12,20-dimethoxy-6,8,16-trimethyl-5,9-bis(oxidanyl)-2-azabicyclo[16.3.1]docosa-1(21),6,15,18(22),19-pentaene-3,11-dione, 1,2-ETHANEDIOL, ADENOSINE MONOPHOSPHATE, ... | | Authors: | Wei, J, Zheng, J, Zhou, J, Kang, Q, Bai, L. | | Deposit date: | 2021-11-17 | | Release date: | 2022-11-16 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.81 Å) | | Cite: | Endowing homodimeric carbamoyltransferase GdmN with iterative functions through structural characterization and mechanistic studies.
Nat Commun, 13, 2022
|
|
7VYJ
 
 | | The structure of GdmN in complex with carbamoyl adenylate intermediate | | Descriptor: | 1,2-ETHANEDIOL, 5'-O-[(S)-(carbamoyloxy)(hydroxy)phosphoryl]adenosine, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Wei, J, Zheng, J, Zhou, J, Kang, Q, Bai, L. | | Deposit date: | 2021-11-14 | | Release date: | 2022-11-16 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Endowing homodimeric carbamoyltransferase GdmN with iterative functions through structural characterization and mechanistic studies.
Nat Commun, 13, 2022
|
|
7VYO
 
 | | The structure of GdmN | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Wei, J, Zheng, J, Zhou, J, Kang, Q, Bai, L. | | Deposit date: | 2021-11-14 | | Release date: | 2022-11-16 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Endowing homodimeric carbamoyltransferase GdmN with iterative functions through structural characterization and mechanistic studies.
Nat Commun, 13, 2022
|
|
7VZZ
 
 | | The structure of GdmN in complex with the natural tetrahedral intermediate, carbamoyl adenylate, and 20-O-methyl-19-chloroproansamitocin | | Descriptor: | (5~{S},6~{E},8~{S},9~{S},12~{R},15~{E})-21-chloranyl-12,20-dimethoxy-6,8,16-trimethyl-5,9-bis(oxidanyl)-2-azabicyclo[16.3.1]docosa-1(21),6,15,18(22),19-pentaene-3,11-dione, 1,2-ETHANEDIOL, 5'-O-[(S)-(carbamoyloxy)(hydroxy)phosphoryl]adenosine, ... | | Authors: | Wei, J, Zheng, J, Zhou, J, Kang, Q, Bai, L. | | Deposit date: | 2021-11-17 | | Release date: | 2022-11-16 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Endowing homodimeric carbamoyltransferase GdmN with iterative functions through structural characterization and mechanistic studies.
Nat Commun, 13, 2022
|
|
7VKT
 
 | | cryo-EM structure of LTB4-bound BLT1 in complex with Gi protein | | Descriptor: | CHOLESTEROL HEMISUCCINATE, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | He, Y, Wang, N. | | Deposit date: | 2021-10-01 | | Release date: | 2022-03-09 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structural basis of leukotriene B4 receptor 1 activation.
Nat Commun, 13, 2022
|
|
8XXV
 
 | | Cryo-EM Structure of the Prostaglandin D2 Receptor 2-indomethacin Coupled to G Protein | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(i) subunit alpha-1, ... | | Authors: | Xu, J, Xu, Y, Wu, C, Xu, H.E. | | Deposit date: | 2024-01-19 | | Release date: | 2024-12-04 | | Last modified: | 2025-02-26 | | Method: | ELECTRON MICROSCOPY (2.33 Å) | | Cite: | Molecular basis of lipid and ligand regulation of prostaglandin receptor DP2.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
8XXU
 
 | | Cryo-EM Structure of the Prostaglandin D2 Receptor 2 Coupled to G Protein | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(i) subunit alpha-1, ... | | Authors: | Xu, J, Xu, Y, Wu, C, Xu, H.E. | | Deposit date: | 2024-01-19 | | Release date: | 2024-12-04 | | Last modified: | 2025-02-26 | | Method: | ELECTRON MICROSCOPY (2.54 Å) | | Cite: | Molecular basis of lipid and ligand regulation of prostaglandin receptor DP2.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
8AHH
 
 | |
8AHG
 
 | |
8AHI
 
 | |
8AHE
 
 | | PAC-FragmentDEL: Photoactivated covalent capture of DNA encoded fragments for hit discovery | | Descriptor: | SULFATE ION, UDP-N-acetylglucosamine 2-epimerase, ~{N},5-dimethyl-1-phenyl-pyrazole-4-sulfonamide | | Authors: | Baker, L.M, Murray, J.B, Hubbard, R.E. | | Deposit date: | 2022-07-21 | | Release date: | 2022-09-14 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.108 Å) | | Cite: | PAC-FragmentDEL - photoactivated covalent capture of DNA-encoded fragments for hit discovery.
Rsc Med Chem, 13, 2022
|
|
8AHF
 
 | | PAC-FragmentDEL: Photoactivated covalent capture of DNA encoded fragments for hit discovery | | Descriptor: | (2~{R},4~{S})-4-[bis(fluoranyl)methoxy]-~{N}-methyl-1-(2~{H}-pyrazolo[4,3-b]pyridin-6-ylcarbonyl)pyrrolidine-2-carboxamide, SULFATE ION, UDP-N-acetylglucosamine 2-epimerase | | Authors: | Baker, L.M, Murray, J.B, Hubbard, R.E. | | Deposit date: | 2022-07-21 | | Release date: | 2022-09-14 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.271 Å) | | Cite: | PAC-FragmentDEL - photoactivated covalent capture of DNA-encoded fragments for hit discovery.
Rsc Med Chem, 13, 2022
|
|
8FQ7
 
 | | Nanobody with WIW inserted in CDR3 loop to Inhibit Growth of Alzheimer's Tau fibrils | | Descriptor: | GLYCEROL, Nanobody with WIW insert in CDR3 loop to target tau fibrils | | Authors: | Abskharon, R, Sawaya, M.R, Cascio, D.C, Eisenberg, D.S. | | Deposit date: | 2023-01-05 | | Release date: | 2023-10-04 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structure-based design of nanobodies that inhibit seeding of Alzheimer's patient-extracted tau fibrils.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
8GAP
 
 | | Structure of LARP7 protein p65-telomerase RNA complex in telomerase | | Descriptor: | Telomerase La-related protein p65, Telomerase RNA, Telomerase associated protein p50, ... | | Authors: | Wang, Y, He, Y, Wang, Y, Yang, Y, Singh, M, Eichhorn, C.D, Zhou, Z.H, Feigon, J. | | Deposit date: | 2023-02-23 | | Release date: | 2023-06-28 | | Last modified: | 2025-06-04 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structure of LARP7 Protein p65-telomerase RNA Complex in Telomerase Revealed by Cryo-EM and NMR.
J.Mol.Biol., 435, 2023
|
|
8XIQ
 
 | | Structure of L796778-SSTR3 G protein complex | | Descriptor: | G-alpha i, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Wang, Y, Xu, Y, Xu, H.E, Zhuang, Y. | | Deposit date: | 2023-12-19 | | Release date: | 2024-07-03 | | Last modified: | 2025-01-15 | | Method: | ELECTRON MICROSCOPY (2.71 Å) | | Cite: | Selective ligand recognition and activation of somatostatin receptors SSTR1 and SSTR3.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
8XIO
 
 | | Structure of L797591-SSTR1 G protein complex | | Descriptor: | (2~{S})-~{N}-[(4~{S})-6-azanyl-2,2,4-trimethyl-hexyl]-3-naphthalen-1-yl-2-[[2-phenylethyl(2-pyridin-2-ylethyl)carbamoyl]amino]propanamide, G-alpha-i, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Wang, Y, Xu, Y, Xu, H.E, Zhuang, Y. | | Deposit date: | 2023-12-19 | | Release date: | 2024-07-03 | | Last modified: | 2025-01-15 | | Method: | ELECTRON MICROSCOPY (2.65 Å) | | Cite: | Selective ligand recognition and activation of somatostatin receptors SSTR1 and SSTR3.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
8XIR
 
 | | Structure of pasireotide-SSTR3 G protein complex | | Descriptor: | G-alpha i, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Wang, Y, Xu, Y, Xu, H.E, Zhuang, Y. | | Deposit date: | 2023-12-19 | | Release date: | 2024-07-03 | | Last modified: | 2025-01-15 | | Method: | ELECTRON MICROSCOPY (2.52 Å) | | Cite: | Selective ligand recognition and activation of somatostatin receptors SSTR1 and SSTR3.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
8XIP
 
 | | Structure of Pasireotide-SSTR1 G protein complex | | Descriptor: | 004-DTR-LYS-TY5-PHE-A1D5E, G-alpha i, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Wang, Y, Xu, Y, Xu, H.E, Zhuang, Y. | | Deposit date: | 2023-12-19 | | Release date: | 2024-07-03 | | Last modified: | 2025-01-15 | | Method: | ELECTRON MICROSCOPY (3.29 Å) | | Cite: | Selective ligand recognition and activation of somatostatin receptors SSTR1 and SSTR3.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
