7WC5
 
 | | Crystal structure of serotonin 2A receptor in complex with psilocin | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 3-[2-(dimethylamino)ethyl]-1~{H}-indol-4-ol, 5-hydroxytryptamine receptor 2A, ... | | Authors: | Cao, D, Yu, J, Wang, H, Luo, Z, Liu, X, He, L, Qi, J, Fan, L, Tang, L, Chen, Z, Li, J, Cheng, J, Wang, S. | | Deposit date: | 2021-12-18 | | Release date: | 2022-01-26 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structure-based discovery of nonhallucinogenic psychedelic analogs.
Science, 375, 2022
|
|
7WC6
 
 | | Crystal structure of serotonin 2A receptor in complex with LSD | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, (8alpha)-N,N-diethyl-6-methyl-9,10-didehydroergoline-8-carboxamide, 5-hydroxytryptamine receptor 2A,Soluble cytochrome b562, ... | | Authors: | Cao, D, Yu, J, Wang, H, Luo, Z, Liu, X, He, L, Qi, J, Fan, L, Tang, L, Chen, Z, Li, J, Cheng, J, Wang, S. | | Deposit date: | 2021-12-18 | | Release date: | 2022-01-26 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure-based discovery of nonhallucinogenic psychedelic analogs.
Science, 375, 2022
|
|
7WC9
 
 | | Crystal structure of serotonin 2A receptor in complex with non-hallucinogenic psychedelic analog | | Descriptor: | (10~{R},15~{S})-12-[3-(2-methoxyphenyl)propyl]-4-methyl-1,4,12-triazatetracyclo[7.6.1.0^{5,16}.0^{10,15}]hexadeca-5(16),6,8-triene, (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 5-hydroxytryptamine receptor 2A,5-hydroxytryptamine receptor 2A,Soluble cytochrome b562, ... | | Authors: | Cao, D, Yu, J, Wang, H, Luo, Z, Liu, X, He, L, Qi, J, Fan, L, Tang, L, Chen, Z, Li, J, Cheng, J, Wang, S. | | Deposit date: | 2021-12-18 | | Release date: | 2022-01-26 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure-based discovery of nonhallucinogenic psychedelic analogs.
Science, 375, 2022
|
|
7WC7
 
 | | Crystal structure of serotonin 2A receptor in complex with lisuride | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 5-hydroxytryptamine receptor 2A,5-hydroxytryptamine receptor 2A,Soluble cytochrome b562, CHOLESTEROL, ... | | Authors: | Cao, D, Yu, J, Wang, H, Luo, Z, Liu, X, He, L, Qi, J, Fan, L, Tang, L, Chen, Z, Li, J, Cheng, J, Wang, S. | | Deposit date: | 2021-12-18 | | Release date: | 2022-01-26 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure-based discovery of nonhallucinogenic psychedelic analogs.
Science, 375, 2022
|
|
7WC8
 
 | | Crystal structure of serotonin 2A receptor in complex with lumateperone | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 1-(4-fluorophenyl)-4-[(10~{R},15~{S})-4-methyl-1,4,12-triazatetracyclo[7.6.1.0^{5,16}.0^{10,15}]hexadeca-5,7,9(16)-trien-12-yl]butan-1-one, 5-hydroxytryptamine receptor 2A,5-hydroxytryptamine receptor 2A,Soluble cytochrome b562, ... | | Authors: | Cao, D, Yu, J, Wang, H, Luo, Z, Liu, X, He, L, Qi, J, Fan, L, Tang, L, Chen, Z, Li, J, Cheng, J, Wang, S. | | Deposit date: | 2021-12-18 | | Release date: | 2022-01-26 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Structure-based discovery of nonhallucinogenic psychedelic analogs.
Science, 375, 2022
|
|
7WC4
 
 | | Crystal structure of serotonin 2A receptor in complex with serotonin | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 5-hydroxytryptamine receptor 2A,5-hydroxytryptamine receptor 2A,5-hydroxytryptamine receptor 2A,Soluble cytochrome b562, CHOLESTEROL, ... | | Authors: | Cao, D, Yu, J, Wang, H, Luo, Z, Liu, X, He, L, Qi, J, Fan, L, Tang, L, Chen, Z, Li, J, Cheng, J, Wang, S. | | Deposit date: | 2021-12-18 | | Release date: | 2022-01-26 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structure-based discovery of nonhallucinogenic psychedelic analogs.
Science, 375, 2022
|
|
7WZB
 
 | | lipopolysaccharide assembly protein LapB (open) | | Descriptor: | Lipopolysaccharide assembly protein B, TRIETHYLENE GLYCOL, ZINC ION | | Authors: | Yan, L, Dong, H, Li, H, Liu, X, Deng, Z, Dong, C, Zhang, Z. | | Deposit date: | 2022-02-17 | | Release date: | 2023-03-01 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | lipopolysaccharide assembly protein LapB (open)
To Be Published
|
|
7Y9V
 
 | | Structure of the auxin exporter PIN1 in Arabidopsis thaliana in the IAA-bound state | | Descriptor: | 1H-INDOL-3-YLACETIC ACID, Auxin efflux carrier component 1, nanobody | | Authors: | Sun, L, Liu, X, Yang, Z, Xia, J. | | Deposit date: | 2022-06-26 | | Release date: | 2022-09-07 | | Last modified: | 2022-09-28 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural insights into auxin recognition and efflux by Arabidopsis PIN1.
Nature, 609, 2022
|
|
7Y9U
 
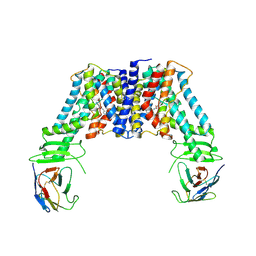 | | Structure of the auxin exporter PIN1 in Arabidopsis thaliana in the NPA-bound state | | Descriptor: | 2-(naphthalen-1-ylcarbamoyl)benzoic acid, Auxin efflux carrier component 1, nanobody | | Authors: | Sun, L, Liu, X, Yang, Z, Xia, J. | | Deposit date: | 2022-06-26 | | Release date: | 2022-09-07 | | Last modified: | 2022-09-28 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural insights into auxin recognition and efflux by Arabidopsis PIN1.
Nature, 609, 2022
|
|
7Y9T
 
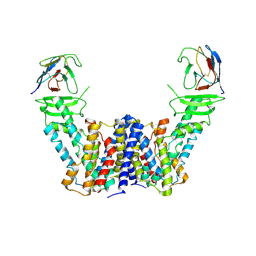 | | Structure of the auxin exporter PIN1 in Arabidopsis thaliana in the apo state | | Descriptor: | Auxin efflux carrier component 1, nanobody | | Authors: | Sun, L, Liu, X, Yang, Z, Xia, J. | | Deposit date: | 2022-06-26 | | Release date: | 2022-09-07 | | Last modified: | 2022-09-28 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural insights into auxin recognition and efflux by Arabidopsis PIN1.
Nature, 609, 2022
|
|
8BAY
 
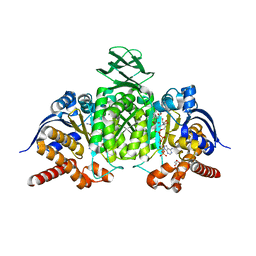 | | Crystal Structure of IDH1 variant R132C S280F in complex with NADPH, Ca2+ and 3-butyl-2-oxoglutarate | | Descriptor: | (R)-3-butyl-2-oxopentanedioic acid, (S)-3-butyl-2-oxopentanedioic acid, CALCIUM ION, ... | | Authors: | Rabe, P, Schofield, C.J, Reinbold, R, Brewitz, L. | | Deposit date: | 2022-10-12 | | Release date: | 2022-11-02 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Natural and synthetic 2-oxoglutarate derivatives are substrates for oncogenic variants of human isocitrate dehydrogenase 1 and 2.
J.Biol.Chem., 299, 2023
|
|
4NX2
 
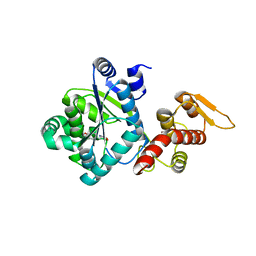 | | Crystal structure of DCYRS complexed with DCY | | Descriptor: | 3,5-dichloro-L-tyrosine, Tyrosine--tRNA ligase | | Authors: | Wang, J, Gong, W, Li, J, Gao, F, Li, H. | | Deposit date: | 2013-12-08 | | Release date: | 2014-09-24 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Significant expansion of fluorescent protein sensing ability through the genetic incorporation of superior photo-induced electron-transfer quenchers.
J.Am.Chem.Soc., 136, 2014
|
|
6L31
 
 | | L1 protein of human papillomavirus 6 | | Descriptor: | Major capsid protein L1 | | Authors: | Li, S.W, Liu, X.L, Gu, Y. | | Deposit date: | 2019-10-07 | | Release date: | 2019-12-25 | | Method: | ELECTRON MICROSCOPY (4.18 Å) | | Cite: | Neutralization sites of human papillomavirus-6 relate to virus attachment and entry phase in viral infection.
Emerg Microbes Infect, 8, 2019
|
|
8XJ4
 
 | | Structure of prostatic acid phosphatase in human semen | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Prostatic acid phosphatase, alpha-D-mannopyranose, ... | | Authors: | Liu, X.Z, Li, J.L, Deng, D, Wang, X. | | Deposit date: | 2023-12-20 | | Release date: | 2024-02-28 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (3.19 Å) | | Cite: | Purification, identification and Cryo-EM structure of prostatic acid phosphatase in human semen.
Biochem.Biophys.Res.Commun., 702, 2024
|
|
2LWF
 
 | | Structure of N-terminal domain of a plant Grx | | Descriptor: | Monothiol glutaredoxin-S16, chloroplastic | | Authors: | Feng, Y. | | Deposit date: | 2012-07-28 | | Release date: | 2013-05-22 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural insights into the N-terminal GIY-YIG endonuclease activity of Arabidopsis glutaredoxin AtGRXS16 in chloroplasts.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
6AE9
 
 | | X-ray structure of the photosystem II phosphatase PBCP | | Descriptor: | (20S)-2,5,8,11,14,17-HEXAMETHYL-3,6,9,12,15,18-HEXAOXAHENICOSANE-1,20-DIOL, MAGNESIUM ION, MANGANESE (II) ION, ... | | Authors: | Liu, X.Y, Chai, J.C, Ou, X.M, Liu, Z.F. | | Deposit date: | 2018-08-03 | | Release date: | 2019-01-02 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Structural Insights into Substrate Selectivity, Catalytic Mechanism, and Redox Regulation of Rice Photosystem II Core Phosphatase.
Mol Plant, 12, 2019
|
|
4ESR
 
 | | Molecular and Structural Characterization of the SH3 Domain of AHI-1 in Regulation of Cellular Resistance of BCR-ABL+ Chronic Myeloid Leukemia Cells to Tyrosine Kinase Inhibitors | | Descriptor: | DI(HYDROXYETHYL)ETHER, Jouberin | | Authors: | Van Petegem, X.F, Liu, P.X, Lobo, P, Jiang, X. | | Deposit date: | 2012-04-23 | | Release date: | 2012-06-06 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | Molecular and structural characterization of the SH3 domain of AHI-1 in regulation of cellular resistance of BCR-ABL(+) chronic myeloid leukemia cells to tyrosine kinase inhibitors.
Proteomics, 12, 2012
|
|
5GT2
 
 | | Crystal Structure and Biochemical Features of dye-decolorizing peroxidase YfeX from Escherichia coli O157 | | Descriptor: | PROTOPORPHYRIN IX CONTAINING FE, Probable deferrochelatase/peroxidase YfeX | | Authors: | Ma, Y.L, Yuan, Z.G, Liu, S, Wang, J.X, Gu, L.C, Liu, X.H. | | Deposit date: | 2016-08-18 | | Release date: | 2017-02-08 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.093 Å) | | Cite: | Crystal structure and biochemical features of dye-decolorizing peroxidase YfeX from Escherichia coli O157 Asp(143) and Arg(232) play divergent roles toward different substrates
Biochem. Biophys. Res. Commun., 484, 2017
|
|
3AWD
 
 | | Crystal structure of gox2181 | | Descriptor: | CADMIUM ION, MAGNESIUM ION, Putative polyol dehydrogenase | | Authors: | Adam Yuan, Y, Yuan, Z. | | Deposit date: | 2011-03-18 | | Release date: | 2012-02-01 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Biochemical and structural analysis of Gox2181, a new member of the SDR superfamily from Gluconobacter oxydans.
Biochem.Biophys.Res.Commun., 415, 2011
|
|
7W26
 
 | | monolignol ferulate transferase | | Descriptor: | Ferulate monolignol transferase | | Authors: | Xi, L, Shuliu, D, Yue, F, Yi, Z. | | Deposit date: | 2021-11-22 | | Release date: | 2022-03-30 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.43 Å) | | Cite: | Crystal structure of the plant feruloyl-coenzyme A monolignol transferase provides insights into the formation of monolignol ferulate conjugates.
Biochem.Biophys.Res.Commun., 594, 2022
|
|
3ZME
 
 | |
6JGF
 
 | |
6JGV
 
 | |
6JNI
 
 | |
7EE5
 
 | | Crystal structure of Neu5Gc bound PltC | | Descriptor: | N-glycolyl-alpha-neuraminic acid, N-glycolyl-alpha-neuraminic acid-(2-3)-beta-D-galactopyranose-(1-4)-alpha-D-glucopyranose, Subtilase cytotoxin subunit B-like protein, ... | | Authors: | Liu, X.Y, Chen, Z, Gao, X. | | Deposit date: | 2021-03-17 | | Release date: | 2021-12-22 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.24 Å) | | Cite: | Molecular Insights into the Assembly and Functional Diversification of Typhoid Toxin.
Mbio, 13, 2022
|
|
