5UEZ
 
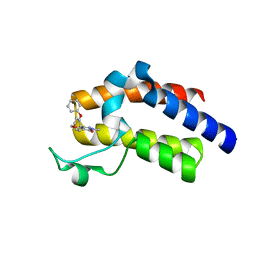 | | BRD4_BD2_A-1342843 | | Descriptor: | 5-methoxy-2-methyl-6-(2-phenoxyphenyl)pyridazin-3(2H)-one, Bromodomain-containing protein 4 | | Authors: | Park, C.H. | | Deposit date: | 2017-01-03 | | Release date: | 2017-05-10 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | Fragment-Based, Structure-Enabled Discovery of Novel Pyridones and Pyridone Macrocycles as Potent Bromodomain and Extra-Terminal Domain (BET) Family Bromodomain Inhibitors.
J. Med. Chem., 60, 2017
|
|
5UF0
 
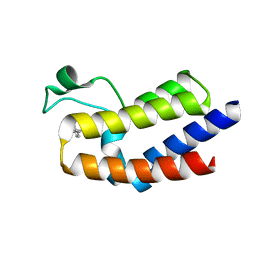 | | BRD4_BD2-A-35165 | | Descriptor: | 2-methyl-5-(methylamino)-6-phenylpyridazin-3(2H)-one, Bromodomain-containing protein 4 | | Authors: | Park, C.H. | | Deposit date: | 2017-01-03 | | Release date: | 2017-06-28 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Fragment-Based, Structure-Enabled Discovery of Novel Pyridones and Pyridone Macrocycles as Potent Bromodomain and Extra-Terminal Domain (BET) Family Bromodomain Inhibitors.
J. Med. Chem., 60, 2017
|
|
5UEW
 
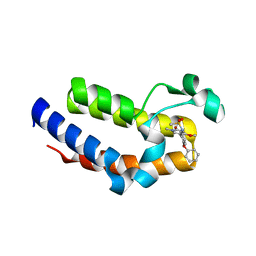 | | BRD2 Bromodomain2 with A-1360579 | | Descriptor: | Bromodomain-containing protein 2, N-[3-(4-methoxy-1-methyl-6-oxo-1,6-dihydropyridin-3-yl)-4-phenoxyphenyl]methanesulfonamide | | Authors: | Park, C.H. | | Deposit date: | 2017-01-03 | | Release date: | 2017-05-10 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Fragment-Based, Structure-Enabled Discovery of Novel Pyridones and Pyridone Macrocycles as Potent Bromodomain and Extra-Terminal Domain (BET) Family Bromodomain Inhibitors.
J. Med. Chem., 60, 2017
|
|
5UEY
 
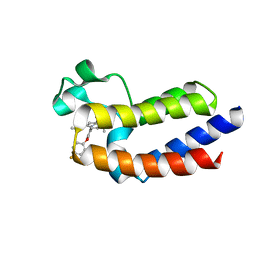 | | BRD4_BD2_A-1412838 | | Descriptor: | 5-[2-(2,4-difluorophenoxy)-5-{[ethyl(dihydroxy)-lambda~4~-sulfanyl]amino}phenyl]-4-ethoxy-1-methylpyridin-2(1H)-one, Bromodomain-containing protein 4 | | Authors: | Park, C.H. | | Deposit date: | 2017-01-03 | | Release date: | 2017-05-10 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.41 Å) | | Cite: | Fragment-Based, Structure-Enabled Discovery of Novel Pyridones and Pyridone Macrocycles as Potent Bromodomain and Extra-Terminal Domain (BET) Family Bromodomain Inhibitors.
J. Med. Chem., 60, 2017
|
|
7YF1
 
 | |
6VWC
 
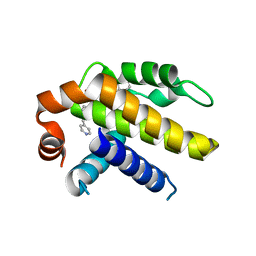 | | Crystal structure of Bcl-xL in complex with tetrahydroisoquinoline-pyridine based inhibitors | | Descriptor: | 6-{8-[(1,3-benzothiazol-2-yl)carbamoyl]-3,4-dihydroisoquinolin-2(1H)-yl}-3-{1-[(pyridin-4-yl)methyl]-1H-pyrazol-4-yl}pyridine-2-carboxylic acid, Bcl-2-like protein 1 | | Authors: | Judge, R.A, Judd, A.S. | | Deposit date: | 2020-02-19 | | Release date: | 2020-10-21 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.604 Å) | | Cite: | Discovery of A-1331852, a First-in-Class, Potent, and Orally-Bioavailable BCL-X L Inhibitor.
Acs Med.Chem.Lett., 11, 2020
|
|
5U1A
 
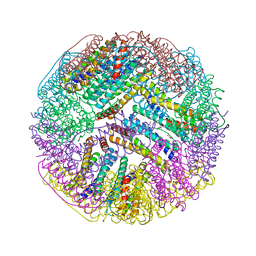 | | Ferritin with Gc MtrE loop 1 inserted at His34 | | Descriptor: | CHLORIDE ION, FE (III) ION, Ferritin,MtrE protein chimera, ... | | Authors: | Wang, S. | | Deposit date: | 2016-11-28 | | Release date: | 2017-10-11 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure-based design of ferritin nanoparticle immunogens displaying antigenic loops of Neisseria gonorrhoeae.
FEBS Open Bio, 7, 2017
|
|
8CEG
 
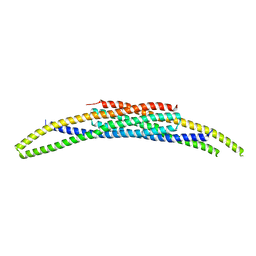 | | BAR domain protein FAM92A1 essential for mitochondrial membrane remodeling | | Descriptor: | CBY1-interacting BAR domain-containing protein 1 | | Authors: | Kajander, T, Fudo, S, Yan, Z, Zhao, H. | | Deposit date: | 2023-02-01 | | Release date: | 2024-02-21 | | Last modified: | 2024-09-04 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Membrane remodeling by FAM92A1 during brain development regulates neuronal morphology, synaptic function, and cognition.
Nat Commun, 15, 2024
|
|
5U1B
 
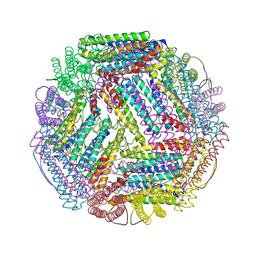 | | Ferritin with Gc MtrE loop2 inserted at the N-terminus | | Descriptor: | MtrE protein,Ferritin chimera | | Authors: | Wang, S. | | Deposit date: | 2016-11-28 | | Release date: | 2017-10-11 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.81 Å) | | Cite: | Structure-based design of ferritin nanoparticle immunogens displaying antigenic loops of Neisseria gonorrhoeae.
FEBS Open Bio, 7, 2017
|
|
6C6Z
 
 | |
8SBD
 
 | | Cryo-EM structure of insulin amyloid-like fibril that is composed of two antiparallel protofilaments | | Descriptor: | Insulin A chain, Insulin B chain | | Authors: | Wang, L.W, Hall, C, Uchikawa, E, Chen, D.L, Choi, E, Zhang, X.W, Bai, X.C. | | Deposit date: | 2023-04-03 | | Release date: | 2023-08-30 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural basis of insulin fibrillation.
Sci Adv, 9, 2023
|
|
6C6Y
 
 | | Crystal structure of Middle-East Respiratory Syndrome (MERS) coronavirus neutralizing antibody JC57-14 isolated from a vaccinated rhesus macaque in complex with MERS Receptor Binding Domain | | Descriptor: | JC57-14 Heavy chain, JC57-14 Light chain, SULFATE ION, ... | | Authors: | Joyce, M.G, Mascola, J.R, Graham, B.S, Kwong, P.D. | | Deposit date: | 2018-01-19 | | Release date: | 2018-03-07 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.32 Å) | | Cite: | Importance of Neutralizing Monoclonal Antibodies Targeting Multiple Antigenic Sites on the Middle East Respiratory Syndrome Coronavirus Spike Glycoprotein To Avoid Neutralization Escape.
J. Virol., 92, 2018
|
|
6C6X
 
 | | Crystal structure of Middle-East Respiratory Syndrome (MERS) coronavirus neutralizing antibody JC57-14 isolated from a vaccinated rhesus macaque. | | Descriptor: | JC57-14 Heavy chain, JC57-14 Light chain | | Authors: | Joyce, M.G, Mascola, J.R, Graham, B.S, Kwong, P.D. | | Deposit date: | 2018-01-19 | | Release date: | 2018-02-28 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Importance of Neutralizing Monoclonal Antibodies Targeting Multiple Antigenic Sites on the Middle East Respiratory Syndrome Coronavirus Spike Glycoprotein To Avoid Neutralization Escape.
J. Virol., 92, 2018
|
|
7MLZ
 
 | | Cryo-EM structure of SARS-CoV-2 spike in complex with neutralizing antibody B1-182.1 that targets the receptor-binding domain | | Descriptor: | B1-182.1 Fab heavy chain, B1-182.1 Fab light chain, Spike protein S1, ... | | Authors: | Zhou, T, Tsybovsky, T, Kwong, P.D. | | Deposit date: | 2021-04-29 | | Release date: | 2021-07-28 | | Last modified: | 2021-08-25 | | Method: | ELECTRON MICROSCOPY (3.71 Å) | | Cite: | Ultrapotent antibodies against diverse and highly transmissible SARS-CoV-2 variants.
Science, 373, 2021
|
|
7MM0
 
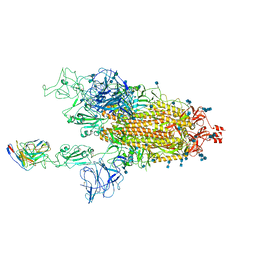 | | Cryo-EM structure of SARS-CoV-2 spike in complex with neutralizing antibody B1-182.1 that targets the receptor-binding domain | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, B1-182.1 Fab heavy chain, ... | | Authors: | Zhou, T, Tsybovsky, T, Kwong, P.D. | | Deposit date: | 2021-04-29 | | Release date: | 2021-07-28 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.15 Å) | | Cite: | Ultrapotent antibodies against diverse and highly transmissible SARS-CoV-2 variants.
Science, 373, 2021
|
|
4ZPV
 
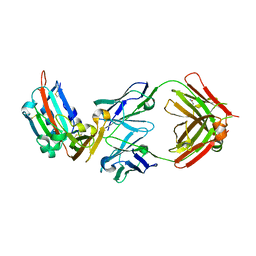 | | Structure of MERS-Coronavirus Spike Receptor-binding Domain (England1 Strain) in Complex with Vaccine-Elicited Murine Neutralizing Antibody D12 (Crystal Form 2) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, D12 Fab Heavy chain, D12 Fab light chain, ... | | Authors: | Joyce, M.G, Mascola, J.R, Graham, B.S, Kwong, P.D. | | Deposit date: | 2015-05-08 | | Release date: | 2015-10-21 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Evaluation of candidate vaccine approaches for MERS-CoV.
Nat Commun, 6, 2015
|
|
4ZPW
 
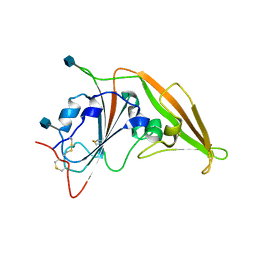 | | Structure of unbound MERS-CoV spike receptor-binding domain (England1 strain). | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, PHOSPHATE ION, Spike glycoprotein | | Authors: | Joyce, M.G, Mascola, J.R, Graham, B.S, Kwong, P.D. | | Deposit date: | 2015-05-08 | | Release date: | 2015-08-12 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (3.023 Å) | | Cite: | Evaluation of candidate vaccine approaches for MERS-CoV.
Nat Commun, 6, 2015
|
|
8JPX
 
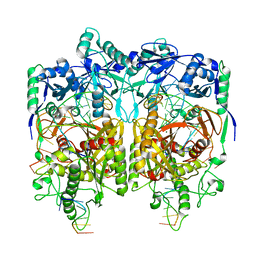 | | Cryo-EM structure of PfAgo-guide DNA-target DNA complex | | Descriptor: | Excess DNA, Guide DNA, MAGNESIUM ION, ... | | Authors: | Zhuang, L. | | Deposit date: | 2023-06-13 | | Release date: | 2024-01-31 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Molecular mechanism for target recognition, dimerization, and activation of Pyrococcus furiosus Argonaute.
Mol.Cell, 84, 2024
|
|
7TB4
 
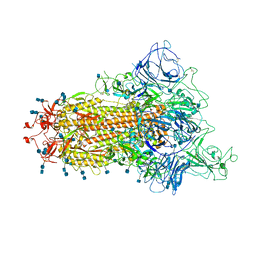 | | Cryo-EM structure of the spike of SARS-CoV-2 Omicron variant of concern | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Surface glycoprotein | | Authors: | Zhou, T, Tsybovsky, T, Kwong, P.D. | | Deposit date: | 2021-12-21 | | Release date: | 2022-01-12 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.29 Å) | | Cite: | Antibodies with potent and broad neutralizing activity against antigenically diverse and highly transmissible SARS-CoV-2 variants.
Biorxiv, 2021
|
|
1BM5
 
 | |
8HBT
 
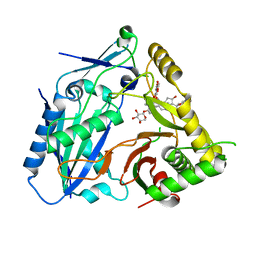 | | AmAT7-3 mutant A310G | | Descriptor: | AmAT7-3-A310G, Astragaloside IV | | Authors: | Wang, L.L. | | Deposit date: | 2022-10-31 | | Release date: | 2023-09-13 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Characterization and structure-based protein engineering of a regiospecific saponin acetyltransferase from Astragalus membranaceus.
Nat Commun, 14, 2023
|
|
8H8I
 
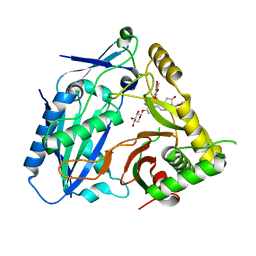 | | Triterpenoid saponin acetyltransferase, AmAT7-3 | | Descriptor: | AmAT7-3, Astragaloside IV | | Authors: | Wang, L.L. | | Deposit date: | 2022-10-22 | | Release date: | 2023-09-13 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Characterization and structure-based protein engineering of a regiospecific saponin acetyltransferase from Astragalus membranaceus.
Nat Commun, 14, 2023
|
|
4ZZ3
 
 | | Human GAR transformylase in complex with GAR and pemetrexed | | Descriptor: | GLYCINAMIDE RIBONUCLEOTIDE, N-{4-[2-(2-amino-4-oxo-4,7-dihydro-3H-pyrrolo[2,3-d]pyrimidin-5-yl)ethyl]benzoyl}-L-glutamic acid, Trifunctional purine biosynthetic protein adenosine-3 | | Authors: | Deis, S.M, Dann III, C.E. | | Deposit date: | 2015-05-22 | | Release date: | 2015-12-16 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.504 Å) | | Cite: | 6-Substituted Pyrrolo[2,3-d]pyrimidine Thienoyl Regioisomers as Targeted Antifolates for Folate Receptor alpha and the Proton-Coupled Folate Transporter in Human Tumors.
J.Med.Chem., 58, 2015
|
|
4ZZ1
 
 | | HUMAN GAR TRANSFORMYLASE IN COMPLEX WITH GAR AND (S)-2-({4-[3-(2-AMINO-4-OXO-4,7-DIHYDRO-3H-PYRROLO[2,3-D]PYRIMIDIN-6-YL)-PROPYL]-THIOPHENE-2-CARBONYL}-AMINO)-PENTANEDIOIC ACID | | Descriptor: | (S)-2-({4-[3-(2-Amino-4-oxo-4,7-dihydro-3H-pyrrolo[2,3-d]pyrimidin-6-yl)-propyl]-thiophene-2-carbonyl}-amino)-pentanedioic acid, GLYCINAMIDE RIBONUCLEOTIDE, Trifunctional purine biosynthetic protein adenosine-3 | | Authors: | Deis, S.M, Dann III, C.E. | | Deposit date: | 2015-05-22 | | Release date: | 2015-12-16 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.351 Å) | | Cite: | 6-Substituted Pyrrolo[2,3-d]pyrimidine Thienoyl Regioisomers as Targeted Antifolates for Folate Receptor alpha and the Proton-Coupled Folate Transporter in Human Tumors.
J.Med.Chem., 58, 2015
|
|
4ZZ2
 
 | | HUMAN GAR TRANSFORMYLASE IN COMPLEX WITH GAR AND (S)-2-({5-[3-(2-AMINO-4-OXO-4,7-DIHYDRO-3H-PYRROLO[2,3-D]PYRIMIDIN-6-YL)-PROPYL]-THIOPHENE-3-CARBONYL}-AMINO)-PENTANEDIOIC ACID | | Descriptor: | (S)-2-({5-[3-(2-Amino-4-oxo-4,7-dihydro-3H-pyrrolo[2,3-d]pyrimidin-6-yl)-propyl]-thiophene-3-carbonyl}-amino)-pentanedioic acid, GLYCINAMIDE RIBONUCLEOTIDE, Trifunctional purine biosynthetic protein adenosine-3 | | Authors: | Deis, S.M, Dann III, C.E. | | Deposit date: | 2015-05-22 | | Release date: | 2015-12-16 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.451 Å) | | Cite: | 6-Substituted Pyrrolo[2,3-d]pyrimidine Thienoyl Regioisomers as Targeted Antifolates for Folate Receptor alpha and the Proton-Coupled Folate Transporter in Human Tumors.
J.Med.Chem., 58, 2015
|
|
