8VJ2
 
 | |
8VR0
 
 | |
8VR1
 
 | |
8W21
 
 | |
8VQZ
 
 | |
8VQW
 
 | |
8W0D
 
 | |
8W0B
 
 | |
8W0C
 
 | |
8W0J
 
 | |
8W0H
 
 | |
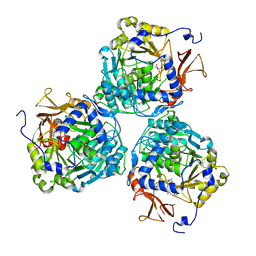
8W0M
 
 | |
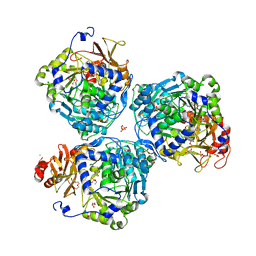
8W0L
 
 | |
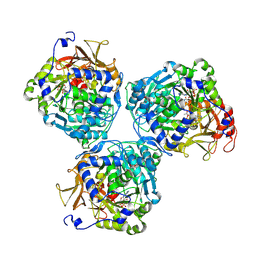
8V4R
 
 | |
5FD2
 
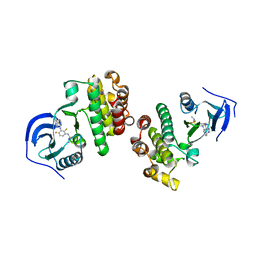 | |
6HHB
 
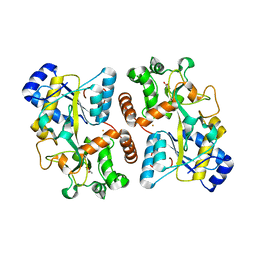 | | Structure of iron bound IbpS from Dickeya dadantii | | Descriptor: | ABC-type Fe3+ transport system, periplasmic component, ACETATE ION, ... | | Authors: | Gueguen-Chaignon, V, Condemine, G, Terradot, L. | | Deposit date: | 2018-08-27 | | Release date: | 2019-09-11 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.80000341 Å) | | Cite: | A secreted metal-binding protein protects necrotrophic phytopathogens from reactive oxygen species.
Nat Commun, 10, 2019
|
|
7VTR
 
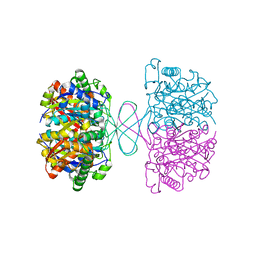 | | 3-ketoacyl-CoA thiolase Tfu_0875 | | Descriptor: | Thiolase | | Authors: | Liu, L.X, Zhou, S.H, Deng, Y. | | Deposit date: | 2021-10-30 | | Release date: | 2022-11-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Rational Design of the Substrate Tunnel of beta-Ketothiolase Reveals a Local Cationic Domain Modulated Rule that Improves the Efficiency of Claisen Condensation
Acs Catalysis, 13, 2023
|
|
7UKL
 
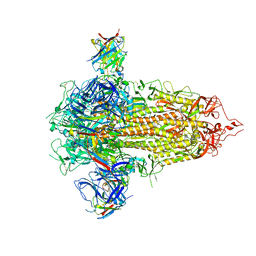 | |
6WJ5
 
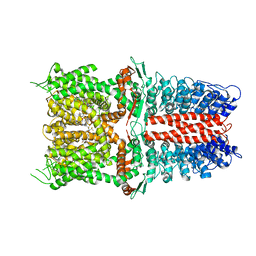 | | Structure of human TRPA1 in complex with inhibitor GDC-0334 | | Descriptor: | (4R,5S)-4-fluoro-1-[(4-fluorophenyl)sulfonyl]-5-methyl-N-({5-(trifluoromethyl)-2-[2-(trifluoromethyl)pyrimidin-5-yl]pyridin-4-yl}methyl)-L-prolinamide, Transient receptor potential cation channel subfamily A member 1 | | Authors: | Rohou, A, Rouge, L, Arthur, C.P, Volgraf, M, Chen, H. | | Deposit date: | 2020-04-11 | | Release date: | 2021-02-17 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | A TRPA1 inhibitor suppresses neurogenic inflammation and airway contraction for asthma treatment.
J.Exp.Med., 218, 2021
|
|
3UUF
 
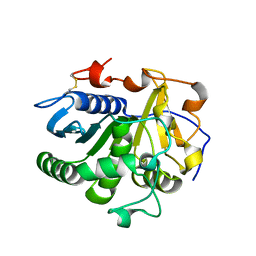 | | Crystal structure of mono- and diacylglycerol lipase from Malassezia globosa | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, LIP1, secretory lipase (Family 3), ... | | Authors: | Xu, T, Xu, J, Hou, S, Liu, J. | | Deposit date: | 2011-11-28 | | Release date: | 2012-04-25 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of a mono- and diacylglycerol lipase from Malassezia globosa reveals a novel lid conformation and insights into the substrate specificity.
J.Struct.Biol., 178, 2012
|
|
2G8X
 
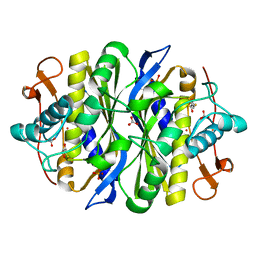 | | Escherichia coli Y209W apoprotein | | Descriptor: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, CARBONATE ION, PHOSPHATE ION, ... | | Authors: | Lee, T.T, Finer-Moore, J.S, Stroud, R.M. | | Deposit date: | 2006-03-03 | | Release date: | 2006-03-14 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | The role of protein dynamics in thymidylate synthase catalysis: Variants of conserved dUMP-binding Tyr-261
TO BE PUBLISHED
|
|
2G8D
 
 | |
2G89
 
 | |
2G86
 
 | |
8B78
 
 | | KRasG12C ligand complex | | Descriptor: | 1-[(4~{a}~{R})-8-(2-chloranyl-6-oxidanyl-phenyl)-7-fluoranyl-9-prop-1-ynyl-1,2,4,4~{a},5,11-hexahydropyrazino[2,1-c][1,4]benzoxazepin-3-yl]propan-1-one, GTPase KRas, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Phillips, C, Breed, J. | | Deposit date: | 2022-09-29 | | Release date: | 2023-07-12 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.11 Å) | | Cite: | Discovery of AZD4747, a Potent and Selective Inhibitor of Mutant GTPase KRAS G12C with Demonstrable CNS Penetration.
J.Med.Chem., 66, 2023
|
|
