7GH1
 
 | | Group deposition SARS-CoV-2 main protease in complex with inhibitors from the COVID Moonshot -- Crystal Structure of SARS-CoV-2 main protease in complex with MAT-POS-2bb0cf2b-1 (Mpro-x12731) | | Descriptor: | (4S)-4-(aminomethyl)-6-chloro-N-(isoquinolin-4-yl)-3,4-dihydro-2H-1-benzopyran-4-carboxamide, 3C-like proteinase, DIMETHYL SULFOXIDE | | Authors: | Fearon, D, Aimon, A, Aschenbrenner, J.C, Balcomb, B.H, Bertram, F.K.R, Brandao-Neto, J, Dias, A, Douangamath, A, Dunnett, L, Godoy, A.S, Gorrie-Stone, T.J, Koekemoer, L, Krojer, T, Lithgo, R.M, Lukacik, P, Marples, P.G, Mikolajek, H, Nelson, E, Owen, C.D, Powell, A.J, Rangel, V.L, Skyner, R, Strain-Damerell, C.M, Thompson, W, Tomlinson, C.W.E, Wild, C, Walsh, M.A, von Delft, F. | | Deposit date: | 2023-08-11 | | Release date: | 2023-11-08 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | Open science discovery of potent noncovalent SARS-CoV-2 main protease inhibitors.
Science, 382, 2023
|
|
7GCT
 
 | | Group deposition SARS-CoV-2 main protease in complex with inhibitors from the COVID Moonshot -- Crystal Structure of SARS-CoV-2 main protease in complex with BRU-LEF-c49414a7-1 (Mpro-x10756) | | Descriptor: | 3C-like proteinase, DIMETHYL SULFOXIDE, N-(4-methylpyridin-3-yl)-2-(3-{[(2R)-4-oxoazetidin-2-yl]oxy}phenyl)acetamide | | Authors: | Fearon, D, Aimon, A, Aschenbrenner, J.C, Balcomb, B.H, Bertram, F.K.R, Brandao-Neto, J, Dias, A, Douangamath, A, Dunnett, L, Godoy, A.S, Gorrie-Stone, T.J, Koekemoer, L, Krojer, T, Lithgo, R.M, Lukacik, P, Marples, P.G, Mikolajek, H, Nelson, E, Owen, C.D, Powell, A.J, Rangel, V.L, Skyner, R, Strain-Damerell, C.M, Thompson, W, Tomlinson, C.W.E, Wild, C, Walsh, M.A, von Delft, F. | | Deposit date: | 2023-08-11 | | Release date: | 2023-11-08 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.857 Å) | | Cite: | Open science discovery of potent noncovalent SARS-CoV-2 main protease inhibitors.
Science, 382, 2023
|
|
7GH7
 
 | | Group deposition SARS-CoV-2 main protease in complex with inhibitors from the COVID Moonshot -- Crystal Structure of SARS-CoV-2 main protease in complex with TRY-UNI-714-22 (Mpro-x2912) | | Descriptor: | (2R)-2-(3-cyanophenyl)-N-(4-methylpyridin-3-yl)propanamide, 3C-like proteinase, DIMETHYL SULFOXIDE | | Authors: | Fearon, D, Aimon, A, Aschenbrenner, J.C, Balcomb, B.H, Bertram, F.K.R, Brandao-Neto, J, Dias, A, Douangamath, A, Dunnett, L, Godoy, A.S, Gorrie-Stone, T.J, Koekemoer, L, Krojer, T, Lithgo, R.M, Lukacik, P, Marples, P.G, Mikolajek, H, Nelson, E, Owen, C.D, Powell, A.J, Rangel, V.L, Skyner, R, Strain-Damerell, C.M, Thompson, W, Tomlinson, C.W.E, Wild, C, Walsh, M.A, von Delft, F. | | Deposit date: | 2023-08-11 | | Release date: | 2023-11-08 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Open science discovery of potent noncovalent SARS-CoV-2 main protease inhibitors.
Science, 382, 2023
|
|
7GD1
 
 | | Group deposition SARS-CoV-2 main protease in complex with inhibitors from the COVID Moonshot -- Crystal Structure of SARS-CoV-2 main protease in complex with BAR-COM-0f94fc3d-58 (Mpro-x10856) | | Descriptor: | (2R)-2-amino-2-(5-bromo-2-methoxyphenyl)-N-(4-methylpyridin-3-yl)acetamide, 3C-like proteinase, DIMETHYL SULFOXIDE | | Authors: | Fearon, D, Aimon, A, Aschenbrenner, J.C, Balcomb, B.H, Bertram, F.K.R, Brandao-Neto, J, Dias, A, Douangamath, A, Dunnett, L, Godoy, A.S, Gorrie-Stone, T.J, Koekemoer, L, Krojer, T, Lithgo, R.M, Lukacik, P, Marples, P.G, Mikolajek, H, Nelson, E, Owen, C.D, Powell, A.J, Rangel, V.L, Skyner, R, Strain-Damerell, C.M, Thompson, W, Tomlinson, C.W.E, Wild, C, Walsh, M.A, von Delft, F. | | Deposit date: | 2023-08-11 | | Release date: | 2023-11-08 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | Open science discovery of potent noncovalent SARS-CoV-2 main protease inhibitors.
Science, 382, 2023
|
|
5EK0
 
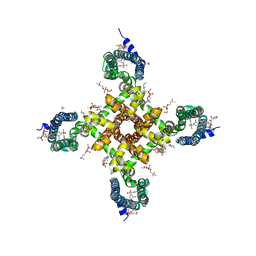 | | Human Nav1.7-VSD4-NavAb in complex with GX-936. | | Descriptor: | 1,2-DIMYRISTOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 3-cyano-4-[2-[2-(1-ethylazetidin-3-yl)pyrazol-3-yl]-4-(trifluoromethyl)phenoxy]-~{N}-(1,2,4-thiadiazol-5-yl)benzenesulfonamide, Chimera of bacterial Ion transport protein and human Sodium channel protein type 9 subunit alpha | | Authors: | Ahuja, S, Mukund, S, Starovasnik, M.A, Koth, C.M, Payandeh, J. | | Deposit date: | 2015-11-03 | | Release date: | 2015-12-23 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.53 Å) | | Cite: | Structural basis of Nav1.7 inhibition by an isoform-selective small-molecule antagonist.
Science, 350, 2015
|
|
6RGV
 
 | |
7RZD
 
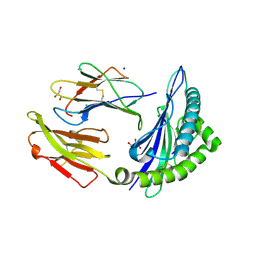 | | CRYSTAL STRUCTURE OF HLA-B*07:02 IN COMPLEX WITH MLL(747-755) PEPTIDE | | Descriptor: | Beta-2-microglobulin, CHLORIDE ION, GLYCEROL, ... | | Authors: | Patskovsky, Y, Patskovska, L, Nyovanie, S, Natarajan, A, Joshi, B, Morin, B, Brittsan, C, Huber, O, Gordon, S, Michelet, X, Schmitzberger, F, Stein, R, Findeis, M, Hurwitz, A, Van Dijk, M, Buell, J, Underwood, D, Krogsgaard, M. | | Deposit date: | 2021-08-27 | | Release date: | 2022-11-02 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Molecular mechanism of phosphopeptide neoantigen immunogenicity.
Nat Commun, 14, 2023
|
|
7S8A
 
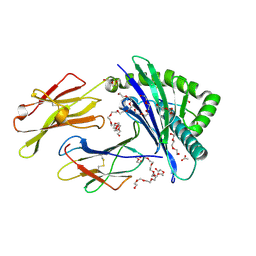 | | STRUCTURE OF HLA-B*07:02 IN COMPLEX WITH MLL(747-755) PHOSPHOPEPTIDE, CUBIC CRYSTAL FORM | | Descriptor: | 3,6,9,12,15,18,21-HEPTAOXATRICOSANE-1,23-DIOL, ACETATE ION, Beta-2-microglobulin, ... | | Authors: | Patskovska, L, Patskovsky, Y, Nyovanie, S, Natarajan, A, Joshi, B, Morin, B, Brittsan, C, Huber, O, Gordon, S, Michelet, X, Schmitzberger, F, Stein, R, Findeis, M, Hurwitz, A, Van Dijk, M, Buell, J, Underwood, D, Krogsgaard, M. | | Deposit date: | 2021-09-17 | | Release date: | 2022-11-02 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Molecular mechanism of phosphopeptide neoantigen immunogenicity.
Nat Commun, 14, 2023
|
|
7S7F
 
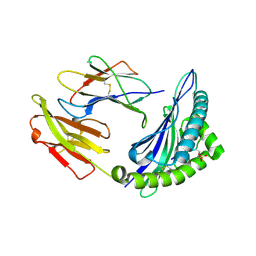 | | STRUCTURE OF HLA-B*07:02 IN COMPLEX WITH DOT1L(998-1006) PHOSPHOPEPTIDE | | Descriptor: | Beta-2-microglobulin, HLA class I histocompatibility antigen, B-7 alpha chain, ... | | Authors: | Patskovska, L, Patskovsky, Y, Nyovanie, S, Natarajan, A, Joshi, B, Morin, B, Brittsan, C, Huber, O, Gordon, S, Michelet, X, Schmitzberger, F, Stein, R, Findeis, M, Hurwitz, A, Van Dijk, M, Buell, J, Underwood, D, Krogsgaard, M. | | Deposit date: | 2021-09-15 | | Release date: | 2022-11-02 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Molecular mechanism of phosphopeptide neoantigen immunogenicity.
Nat Commun, 14, 2023
|
|
7S7E
 
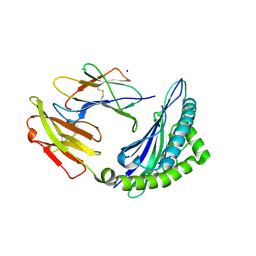 | | STRUCTURE OF HLA-B*07:02 IN COMPLEX WITH DOT1L(998-1006) PEPTIDE | | Descriptor: | Beta-2-microglobulin, HLA class I histocompatibility antigen, B-7 alpha chain, ... | | Authors: | Patskovska, L, Patskovsky, Y, Nyovanie, S, Natarajan, A, Joshi, B, Morin, B, Brittsan, C, Huber, O, Gordon, S, Michelet, X, Schmitzberger, F, Stein, R, Findeis, M, Hurwitz, A, Van Dijk, M, Buell, J, Underwood, D, Krogsgaard, M. | | Deposit date: | 2021-09-15 | | Release date: | 2022-11-02 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Molecular mechanism of phosphopeptide neoantigen immunogenicity.
Nat Commun, 14, 2023
|
|
7S79
 
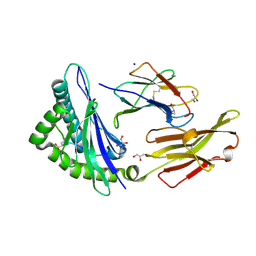 | | STRUCTURE OF HLA-B*07:02 IN COMPLEX WITH SYNTHETIC PHOSPHONO-MLL PEPTIDE ANALOG | | Descriptor: | Beta-2-microglobulin, GLYCEROL, HLA class I histocompatibility antigen, ... | | Authors: | Patskovska, L, Patskovsky, Y, Nyovanie, S, Natarajan, A, Joshi, B, Morin, B, Brittsan, C, Huber, O, Gordon, S, Michelet, X, Schmitzberger, F, Stein, R, Findeis, M, Hurwitz, A, Van Dijk, M, Buell, J, Underwood, D, Krogsgaard, M. | | Deposit date: | 2021-09-15 | | Release date: | 2022-11-02 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | Molecular mechanism of phosphopeptide neoantigen immunogenicity.
Nat Commun, 14, 2023
|
|
7S8I
 
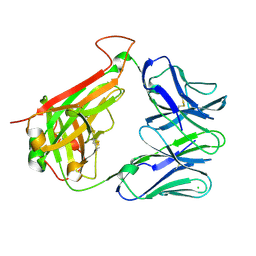 | | PHOSPHOPEPTIDE-SPECIFIC LC13 TCR, MONOCLINIC CRYSTAL FORM | | Descriptor: | CHLORIDE ION, TRAV27_LC13 TCR ALPHA CHAIN, TRBV27_LC13 TCR BETA CHAIN | | Authors: | Patskovsky, Y, Nyovanie, S, Patskovska, L, Natarajan, A, Joshi, B, Morin, B, Brittsan, C, Huber, O, Gordon, S, Michelet, X, Schmitzberger, F, Stein, R, Findeis, M, Hurwitz, A, Van Dijk, M, Buell, J, Underwood, D, Krogsgaard, M. | | Deposit date: | 2021-09-17 | | Release date: | 2022-11-02 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Molecular mechanism of phosphopeptide neoantigen immunogenicity.
Nat Commun, 14, 2023
|
|
7S8J
 
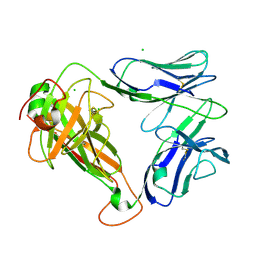 | | PHOSPHOPEPTIDE-SPECIFIC LC13 TCR, ORTHORHOMBIC CRYSTAL FORM | | Descriptor: | CHLORIDE ION, TRAV27_LC13 TCR ALPHA CHAIN, TRBV27_LC13 TCR BETA CHAIN | | Authors: | Patskovska, L, Patskovsky, Y, Nyovanie, S, Natarajan, A, Joshi, B, Morin, B, Brittsan, C, Huber, O, Gordon, S, Michelet, X, Schmitzberger, F, Stein, R, Findeis, M, Hurwitz, A, Van Dijk, M, Buell, J, Underwood, D, Krogsgaard, M. | | Deposit date: | 2021-09-17 | | Release date: | 2022-11-02 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Molecular mechanism of phosphopeptide neoantigen immunogenicity.
Nat Commun, 14, 2023
|
|
7S7D
 
 | | STRUCTURE OF HLA-B*07:02 IN COMPLEX WITH SYNTHETIC SULFO-MLL PEPTIDE ANALOG | | Descriptor: | Beta-2-microglobulin, GLYCEROL, HLA class I histocompatibility antigen, ... | | Authors: | Patskovska, L, Patskovsky, Y, Nyovanie, S, Natarajan, A, Joshi, B, Morin, B, Brittsan, C, Huber, O, Gordon, S, Michelet, X, Schmitzberger, F, Stein, R, Findeis, M, Hurwitz, A, Van Dijk, M, Buell, J, Underwood, D, Krogsgaard, M. | | Deposit date: | 2021-09-15 | | Release date: | 2022-11-02 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | Molecular mechanism of phosphopeptide neoantigen immunogenicity.
Nat Commun, 14, 2023
|
|
7S8E
 
 | | STRUCTURE OF HLA-B*07:02 IN COMPLEX WITH MLL(747-755) PHOSPHOPEPTIDE AND BOUND GLYCEROL | | Descriptor: | Beta-2-microglobulin, GLYCEROL, HLA class I histocompatibility antigen, ... | | Authors: | Patskovsky, Y, Nyovanie, S, Patskovska, L, Natarajan, A, Joshi, B, Morin, B, Brittsan, C, Huber, O, Gordon, S, Michelet, X, Schmitzberger, F, Stein, R, Findeis, M, Hurwitz, A, Van Dijk, M, Buell, J, Underwood, D, Krogsgaard, M. | | Deposit date: | 2021-09-17 | | Release date: | 2022-11-02 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Molecular mechanism of phosphopeptide neoantigen immunogenicity.
Nat Commun, 14, 2023
|
|
7RZJ
 
 | | CRYSTAL STRUCTURE OF HLA-B*07:02 IN COMPLEX WITH MLL(747-755) PHOSPHOPEPTIDE | | Descriptor: | Beta-2-microglobulin, HLA class I histocompatibility antigen, B-7 alpha chain, ... | | Authors: | Patskovsky, Y, Patskovska, L, Nyovanie, S, Natarajan, A, Joshi, B, Morin, B, Brittsan, C, Huber, O, Gordon, S, Michelet, X, Schmitzberger, F, Stein, R, Findeis, M, Hurwitz, A, Van Dijk, M, Buell, J, Underwood, D, Krogsgaard, M. | | Deposit date: | 2021-08-27 | | Release date: | 2022-11-02 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Molecular mechanism of phosphopeptide neoantigen immunogenicity.
Nat Commun, 14, 2023
|
|
7S8F
 
 | | STRUCTURE OF HLA-B*07:02 IN COMPLEX WITH MLL(747-755) PEPTIDE AND BOUND GLYCEROL | | Descriptor: | Beta-2-microglobulin, GLYCEROL, HLA class I histocompatibility antigen, ... | | Authors: | Patskovsky, Y, Nyovanie, S, Patskovska, L, Natarajan, A, Joshi, B, Morin, B, Brittsan, C, Huber, O, Gordon, S, Michelet, X, Schmitzberger, F, Stein, R, Findeis, M, Hurwitz, A, Van Dijk, M, Buell, J, Underwood, D, Krogsgaard, M. | | Deposit date: | 2021-09-17 | | Release date: | 2022-11-02 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Molecular mechanism of phosphopeptide neoantigen immunogenicity.
Nat Commun, 14, 2023
|
|
4D4B
 
 | | The catalytic domain, BcGH76, of Bacillus circulans Aman6 in complex with MSMSMe | | Descriptor: | 1,2-ETHANEDIOL, ALPHA-1,6-MANNANASE, alpha-D-mannopyranose-(1-6)-methyl 1,6-dithio-alpha-D-mannopyranoside | | Authors: | Thompson, A.J, Speciale, G, Iglesias-Fernandez, J, Hakki, Z, Belz, T, Cartmell, A, Spears, R.J, Stepper, J, Gilbert, H.J, Rovira, C, Williams, S.J, Davies, G.J. | | Deposit date: | 2014-10-27 | | Release date: | 2015-03-25 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Evidence for a Boat Conformation at the Transition State of Gh76 Alpha-1,6-Mannanases- Key Enzymes in Bacterial and Fungal Mannoprotein Metabolism
Angew.Chem.Int.Ed.Engl., 54, 2015
|
|
4D4C
 
 | | The catalytic domain, BcGH76, of Bacillus circulans Aman6 in complex with 1,6-ManDMJ | | Descriptor: | 1,2-ETHANEDIOL, 1-DEOXYMANNOJIRIMYCIN, ALPHA-1,6-MANNANASE, ... | | Authors: | Thompson, A.J, Speciale, G, Iglesias-Fernandez, J, Hakki, Z, Belz, T, Cartmell, A, Spears, R.J, Stepper, J, Gilbert, H.J, Rovira, C, Williams, S.J, Davies, G.J. | | Deposit date: | 2014-10-27 | | Release date: | 2015-03-25 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Evidence for a Boat Conformation at the Transition State of Gh76 Alpha-1,6-Mannanases- Key Enzymes in Bacterial and Fungal Mannoprotein Metabolism
Angew.Chem.Int.Ed.Engl., 54, 2015
|
|
4D4D
 
 | | The catalytic domain, BcGH76, of Bacillus circulans Aman6 in complex with 1,6-ManIFG | | Descriptor: | 1,2-ETHANEDIOL, 5-HYDROXYMETHYL-3,4-DIHYDROXYPIPERIDINE, ALPHA-1,6-MANNANASE, ... | | Authors: | Thompson, A.J, Speciale, G, Iglesias-Fernandez, J, Hakki, Z, Belz, T, Cartmell, A, Spears, R.J, Stepper, J, Gilbert, H.J, Rovira, C, Williams, S.J, Davies, G.J. | | Deposit date: | 2014-10-27 | | Release date: | 2015-03-25 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Evidence for a Boat Conformation at the Transition State of Gh76 Alpha-1,6-Mannanases- Key Enzymes in Bacterial and Fungal Mannoprotein Metabolism
Angew.Chem.Int.Ed.Engl., 54, 2015
|
|
4D4A
 
 | | Structure of the catalytic domain (BcGH76) of the Bacillus circulans GH76 alpha mannanase, Aman6. | | Descriptor: | 1,2-ETHANEDIOL, ALPHA-1,6-MANNANASE | | Authors: | Thompson, A.J, Speciale, G, Iglesias-Fernandez, J, Hakki, Z, Belz, T, Cartmell, A, Spears, R.J, Stepper, J, Gilbert, H.J, Rovira, C, Williams, S.J, Davies, G.J. | | Deposit date: | 2014-10-27 | | Release date: | 2015-03-25 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Evidence for a Boat Conformation at the Transition State of Gh76 Alpha-1,6-Mannanases- Key Enzymes in Bacterial and Fungal Mannoprotein Metabolism
Angew.Chem.Int.Ed.Engl., 54, 2015
|
|
