7VVS
 
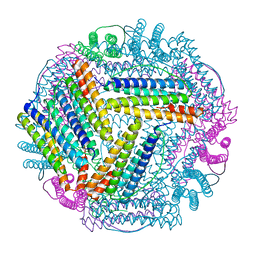 | | PLL9 induced TmFtn nanocage | | Descriptor: | FE (III) ION, Ferritin | | Authors: | Zhao, G, Zhang, X. | | Deposit date: | 2021-11-08 | | Release date: | 2022-11-09 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | PLL9 induced TmFtn nanocage
To Be Published
|
|
3KV6
 
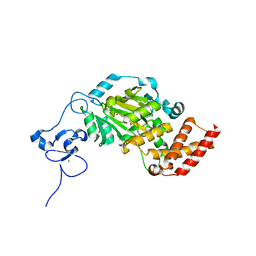 | | Structure of KIAA1718, human Jumonji demethylase, in complex with alpha-ketoglutarate | | Descriptor: | 2-OXOGLUTARIC ACID, FE (II) ION, JmjC domain-containing histone demethylation protein 1D, ... | | Authors: | Horton, J.R, Upadhyay, A.K, Qi, H.H, Zhang, X, Shi, Y, Cheng, X. | | Deposit date: | 2009-11-29 | | Release date: | 2009-12-22 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.89 Å) | | Cite: | Enzymatic and structural insights for substrate specificity of a family of jumonji histone lysine demethylases.
Nat.Struct.Mol.Biol., 17, 2010
|
|
7YD1
 
 | |
7YD0
 
 | |
7YCY
 
 | |
7YCZ
 
 | |
3KV5
 
 | | Structure of KIAA1718, human Jumonji demethylase, in complex with N-oxalylglycine | | Descriptor: | FE (II) ION, JmjC domain-containing histone demethylation protein 1D, N-OXALYLGLYCINE, ... | | Authors: | Horton, J.R, Upadhyay, A.K, Qi, H.H, Zhang, X, Shi, Y, Cheng, X. | | Deposit date: | 2009-11-29 | | Release date: | 2009-12-22 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | Enzymatic and structural insights for substrate specificity of a family of jumonji histone lysine demethylases.
Nat.Struct.Mol.Biol., 17, 2010
|
|
3KVA
 
 | | Structure of KIAA1718 Jumonji domain in complex with alpha-ketoglutarate | | Descriptor: | 2-OXOGLUTARIC ACID, FE (II) ION, JmjC domain-containing histone demethylation protein 1D, ... | | Authors: | Horton, J.R, Upadhyay, A.K, Qi, H.H, Zhang, X, Shi, Y, Cheng, X. | | Deposit date: | 2009-11-29 | | Release date: | 2009-12-22 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.79 Å) | | Cite: | Enzymatic and structural insights for substrate specificity of a family of jumonji histone lysine demethylases.
Nat.Struct.Mol.Biol., 17, 2010
|
|
3KV4
 
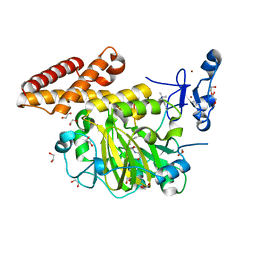 | | Structure of PHF8 in complex with histone H3 | | Descriptor: | 1,2-ETHANEDIOL, FE (II) ION, Histone H3-like, ... | | Authors: | Horton, J.R, Upadhyay, A.K, Qi, H.H, Zhang, X, Shi, Y, Cheng, X. | | Deposit date: | 2009-11-29 | | Release date: | 2009-12-22 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Enzymatic and structural insights for substrate specificity of a family of jumonji histone lysine demethylases.
Nat.Struct.Mol.Biol., 17, 2010
|
|
2DHR
 
 | | Whole cytosolic region of ATP-dependent metalloprotease FtsH (G399L) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, FtsH | | Authors: | Suno, R, Niwa, H, Tsuchiya, D, Zhang, X, Yoshida, M, Morikawa, K. | | Deposit date: | 2006-03-24 | | Release date: | 2006-06-27 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (3.9 Å) | | Cite: | Structure of the Whole Cytosolic Region of ATP-Dependent Protease FtsH
Mol.Cell, 22, 2006
|
|
2DI4
 
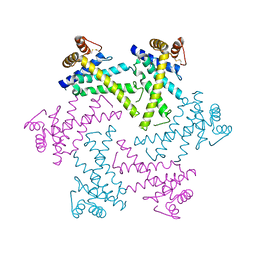 | | Crystal structure of the FtsH protease domain | | Descriptor: | Cell division protein ftsH homolog, MERCURY (II) ION | | Authors: | Suno, R, Niwa, H, Tsuchiya, D, Zhang, X, Yoshida, M, Morikawa, K. | | Deposit date: | 2006-03-28 | | Release date: | 2006-06-27 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.79 Å) | | Cite: | Structure of the Whole Cytosolic Region of ATP-Dependent Protease FtsH
Mol.Cell, 22, 2006
|
|
7QTQ
 
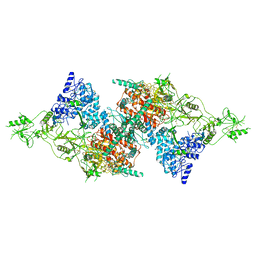 | | Structure of Native, iodinated bovine thyroglobulin solved on strepavidin affinity grids. | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Thyroglobulin, ... | | Authors: | Marechal, N, Weitz, J.C, Serrano, B.P, Zhang, X. | | Deposit date: | 2022-01-15 | | Release date: | 2022-05-04 | | Last modified: | 2023-11-15 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Formation of thyroid hormone revealed by a cryo-EM structure of native bovine thyroglobulin.
Nat Commun, 13, 2022
|
|
3JBL
 
 | | Cryo-EM Structure of the Activated NAIP2/NLRC4 Inflammasome Reveals Nucleated Polymerization | | Descriptor: | NLR family CARD domain-containing protein 4 | | Authors: | Zhang, L, Chen, S, Ruan, J, Wu, J, Tong, A.B, Yin, Q, Li, Y, David, L, Lu, A, Wang, W.L, Marks, C, Ouyang, Q, Zhang, X, Mao, Y, Wu, H. | | Deposit date: | 2015-09-05 | | Release date: | 2015-10-21 | | Last modified: | 2018-07-18 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | Cryo-EM structure of the activated NAIP2-NLRC4 inflammasome reveals nucleated polymerization.
Science, 350, 2015
|
|
2C98
 
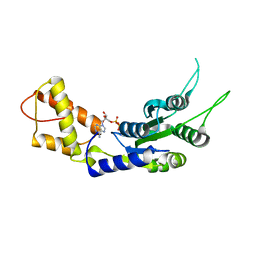 | | Structural basis of the nucleotide driven conformational changes in the AAA domain of transcription activator PspF | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, PSP OPERON TRANSCRIPTIONAL ACTIVATOR | | Authors: | Rappas, M, Schumacher, J, Niwa, H, Buck, M, Zhang, X. | | Deposit date: | 2005-12-09 | | Release date: | 2006-02-01 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural Basis of the Nucleotide Driven Conformational Changes in the Aaa(+) Domain of Transcription Activator Pspf.
J.Mol.Biol., 357, 2006
|
|
2C99
 
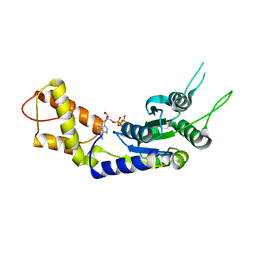 | | Structural basis of the nucleotide driven conformational changes in the AAA domain of transcription activator PspF | | Descriptor: | PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, PSP OPERON TRANSCRIPTIONAL ACTIVATOR | | Authors: | Rappas, M, Schumacher, J, Niwa, H, Buck, M, Zhang, X. | | Deposit date: | 2005-12-09 | | Release date: | 2006-02-01 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural Basis of the Nucleotide Driven Conformational Changes in the Aaa(+) Domain of Transcription Activator Pspf.
J.Mol.Biol., 357, 2006
|
|
2C96
 
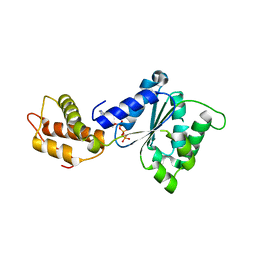 | | Structural basis of the nucleotide driven conformational changes in the AAA domain of transcription activator PspF | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, PSP OPERON TRANSCRIPTIONAL ACTIVATOR | | Authors: | Rappas, M, Schumacher, J, Niwa, H, Buck, M, Zhang, X. | | Deposit date: | 2005-12-09 | | Release date: | 2006-02-01 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural Basis of the Nucleotide Driven Conformational Changes in the Aaa(+) Domain of Transcription Activator Pspf.
J.Mol.Biol., 357, 2006
|
|
2C9C
 
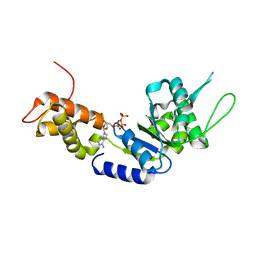 | | Structural basis of the nucleotide driven conformational changes in the AAA domain of transcription activator PspF | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, PSP OPERON TRANSCRIPTIONAL ACTIVATOR | | Authors: | Rappas, M, Schumacher, J, Niwa, H, Buck, M, Zhang, X. | | Deposit date: | 2005-12-09 | | Release date: | 2006-02-01 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural Basis of the Nucleotide Driven Conformational Changes in the Aaa(+) Domain of Transcription Activator Pspf.
J.Mol.Biol., 357, 2006
|
|
2GIC
 
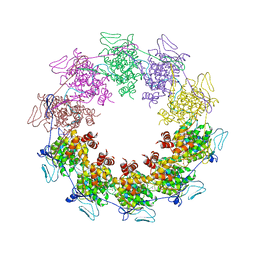 | | Crystal Structure of a vesicular stomatitis virus nucleocapsid-RNA complex | | Descriptor: | 45-MER, Nucleocapsid protein, URANYL (VI) ION | | Authors: | Green, T.J, Zhang, X, Wertz, G.W, Luo, M. | | Deposit date: | 2006-03-28 | | Release date: | 2006-08-22 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.92 Å) | | Cite: | Structure of the vesicular stomatitis virus nucleoprotein-RNA complex unveils how the RNA is sequestered
Science, 313, 2006
|
|
3LOI
 
 | | Crystal structures of Cupin superfamily BbDUF985 from Branchiostoma belcheri tsingtauense in the apo and GDP-bound forms | | Descriptor: | Putative uncharacterized protein | | Authors: | Zou, C.Z, Du, Y, He, Y.-X, Saren, G, Zhang, X, Chen, Y, Zhang, S.-C. | | Deposit date: | 2010-02-04 | | Release date: | 2010-06-02 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structures of the apo and GDP-bound forms of a cupin-like protein BbDUF985 from Branchiostoma belcheri tsingtauense
Proteins, 2010
|
|
3LNL
 
 | | Crystal structure of Staphylococcus aureus protein SA1388 | | Descriptor: | 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, UPF0135 protein SA1388, ZINC ION | | Authors: | Singh, K.S, Chruszcz, M, Zhang, X, Minor, W, Zhang, H. | | Deposit date: | 2010-02-02 | | Release date: | 2010-03-16 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of a Conserved Hypothetical Protein Sa1388 from S. aureus Reveals a Capped Hexameric Toroid with Two Pii Domain Lids and a Dinuclear Metal Center.
Bmc Struct.Biol., 6, 2006
|
|
2GZY
 
 | |
2H3B
 
 | | Crystal Structure of Mouse Nicotinamide Phosphoribosyltransferase/Visfatin/Pre-B Cell Colony Enhancing Factor 1 | | Descriptor: | Nicotinamide phosphoribosyltransferase, SULFATE ION | | Authors: | Wang, T, Zhang, X, Bheda, P, Revollo, J.R, Imai, S.I, Wolberger, C. | | Deposit date: | 2006-05-22 | | Release date: | 2006-06-20 | | Last modified: | 2021-10-20 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structure of Nampt/PBEF/visfatin, a mammalian NAD(+) biosynthetic enzyme.
Nat.Struct.Mol.Biol., 13, 2006
|
|
2GZZ
 
 | |
2H3D
 
 | | Crystal Structure of Mouse Nicotinamide Phosphoribosyltransferase/Visfatin/Pre-B Cell Colony Enhancing Factor in Complex with Nicotinamide Mononuleotide | | Descriptor: | BETA-NICOTINAMIDE RIBOSE MONOPHOSPHATE, Nicotinamide phosphoribosyltransferase | | Authors: | Wang, T, Zhang, X, Bheda, P, Revollo, J.R, Imai, S.I, Wolberger, C. | | Deposit date: | 2006-05-22 | | Release date: | 2006-06-20 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure of Nampt/PBEF/visfatin, a mammalian NAD(+) biosynthetic enzyme.
Nat.Struct.Mol.Biol., 13, 2006
|
|
6O0W
 
 | | Crystal structure of the TIR domain from the grapevine disease resistance protein RUN1 in complex with NADP+ and Bis-Tris | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ADENOSINE-2'-5'-DIPHOSPHATE, TIR-NB-LRR type resistance protein RUN1 | | Authors: | Horsefield, S, Burdett, H, Zhang, X, Manik, M.K, Shi, Y, Chen, J, Tiancong, Q, Gilley, J, Lai, J, Gu, W, Rank, M, Casey, L, Ericsson, D.J, Foley, G, Hughes, R.O, Bosanac, T, von Itzstein, M, Rathjen, J.P, Nanson, J.D, Boden, M, Dry, I.B, Williams, S.J, Staskawicz, B.J, Coleman, M.P, Ve, T, Dodds, P.N, Kobe, B. | | Deposit date: | 2019-02-17 | | Release date: | 2019-09-04 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | NAD+cleavage activity by animal and plant TIR domains in cell death pathways.
Science, 365, 2019
|
|
