7EKE
 
 | | Structure of SARS-CoV-2 spike receptor-binding domain F486L mutation complexed with human ACE2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, Spike protein S1, ... | | Authors: | Han, P.C, Su, C, Zhang, Y.F, Qi, J.X, Gao, G.F. | | Deposit date: | 2021-04-05 | | Release date: | 2021-11-03 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Molecular insights into receptor binding of recent emerging SARS-CoV-2 variants.
Nat Commun, 12, 2021
|
|
7EKG
 
 | | Structure of SARS-CoV-2 Beta variant spike receptor-binding domain complexed with human ACE2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, ... | | Authors: | Han, P.C, Su, C, Zhang, Y.F, Qi, J.X, Gao, G.F. | | Deposit date: | 2021-04-05 | | Release date: | 2021-11-03 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.63 Å) | | Cite: | Molecular insights into receptor binding of recent emerging SARS-CoV-2 variants.
Nat Commun, 12, 2021
|
|
7EKH
 
 | | Structure of SARS-CoV-2 spike receptor-binding domain Y453F mutation complexed with human ACE2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, Spike protein S1, ... | | Authors: | Han, P.C, Su, C, Zhang, Y.F, Qi, J.X, Gao, G.F. | | Deposit date: | 2021-04-05 | | Release date: | 2021-11-03 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Molecular insights into receptor binding of recent emerging SARS-CoV-2 variants.
Nat Commun, 12, 2021
|
|
7EKF
 
 | | Structure of SARS-CoV-2 Alpha variant spike receptor-binding domain complexed with human ACE2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, ... | | Authors: | Han, P.C, Su, C, Zhang, Y.F, Qi, J.X, Gao, G.F. | | Deposit date: | 2021-04-05 | | Release date: | 2021-11-03 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Molecular insights into receptor binding of recent emerging SARS-CoV-2 variants.
Nat Commun, 12, 2021
|
|
7EKC
 
 | | Structure of SARS-CoV-2 Gamma variant spike receptor-binding domain complexed with human ACE2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, ... | | Authors: | Han, P.C, Su, C, Zhang, Y.F, Qi, J.X, Gao, G.F. | | Deposit date: | 2021-04-05 | | Release date: | 2021-11-03 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Molecular insights into receptor binding of recent emerging SARS-CoV-2 variants.
Nat Commun, 12, 2021
|
|
7BZU
 
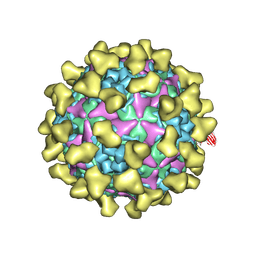 | | Cryo-EM structure of mature Coxsackievirus A10 in complex with KRM1 at pH 5.5 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Capsid protein VP1, Capsid protein VP2, ... | | Authors: | Cui, Y, Peng, R, Song, H, Tong, Z, Gao, G.F, Qi, J. | | Deposit date: | 2020-04-28 | | Release date: | 2020-07-22 | | Last modified: | 2025-06-18 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Molecular basis of Coxsackievirus A10 entry using the two-in-one attachment and uncoating receptor KRM1.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
7E8O
 
 | |
7E8K
 
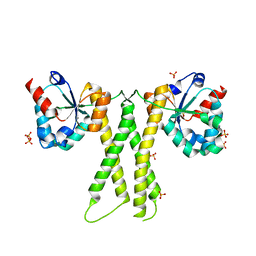 | |
7C4Y
 
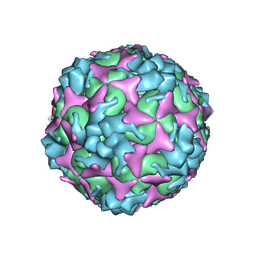 | | Cryo-EM structure of empty Coxsackievirus A10 at pH 7.4 | | Descriptor: | Capsid protein VP1, Capsid protein VP2, Capsid protein VP3 | | Authors: | Cui, Y, Peng, R, Song, H, Tong, Z, Gao, G.F, Qi, J. | | Deposit date: | 2020-05-18 | | Release date: | 2020-07-22 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Molecular basis of Coxsackievirus A10 entry using the two-in-one attachment and uncoating receptor KRM1.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
7E8J
 
 | |
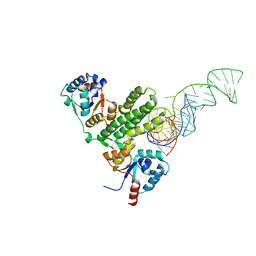
8IR4
 
 | | Crystal structure of the SLF1 BRCT domain in complex with a Rad18 peptide containing pS442 | | Descriptor: | CHLORIDE ION, ISOPROPYL ALCOHOL, MAGNESIUM ION, ... | | Authors: | Xiang, S, Huang, W, Qiu, F. | | Deposit date: | 2023-03-17 | | Release date: | 2023-11-15 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Structural insights into Rad18 targeting by the SLF1 BRCT domains.
J.Biol.Chem., 299, 2023
|
|
8IR2
 
 | | Crystal structure of the SLF1 BRCT domain in complex with a Rad18 peptide containing pS442 and pS444 | | Descriptor: | CHLORIDE ION, ISOPROPYL ALCOHOL, MAGNESIUM ION, ... | | Authors: | Xiang, S, Huang, W, Qiu, F. | | Deposit date: | 2023-03-17 | | Release date: | 2023-11-15 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural insights into Rad18 targeting by the SLF1 BRCT domains.
J.Biol.Chem., 299, 2023
|
|
8IJ1
 
 | | Protomer 1 and 2 of the asymmetry trimer of the Cul2-Rbx1-EloBC-FEM1B ubiquitin ligase complex | | Descriptor: | Cullin-2, E3 ubiquitin-protein ligase RBX1, Elongin-B, ... | | Authors: | Dai, Z, Liang, L, Yin, Y.X. | | Deposit date: | 2023-02-24 | | Release date: | 2024-02-28 | | Last modified: | 2024-04-03 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Structural insights into the ubiquitylation strategy of the oligomeric CRL2 FEM1B E3 ubiquitin ligase.
Embo J., 43, 2024
|
|
8JOP
 
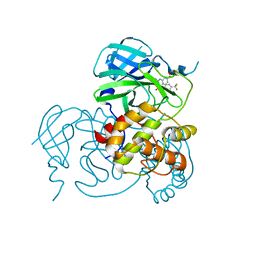 | | Crystal structure of the SARS-CoV-2 main protease in complex with 11a | | Descriptor: | 3C-like proteinase nsp5, methyl (6~{R})-5-ethanoyl-7-oxidanylidene-6-[4-(trifluoromethyl)phenyl]-8,9,10,11-tetrahydro-6~{H}-benzo[b][1,4]benzodiazepine-2-carboxylate | | Authors: | Zeng, R, Liu, Y.Z, Wang, F.L, Yang, S.Y, Lei, J. | | Deposit date: | 2023-06-08 | | Release date: | 2023-08-16 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Discovery of benzodiazepine derivatives as a new class of covalent inhibitors of SARS-CoV-2 main protease.
Bioorg.Med.Chem.Lett., 92, 2023
|
|
8JSL
 
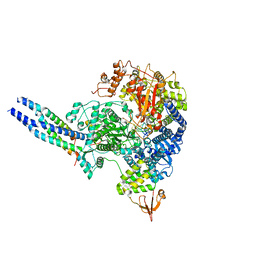 | | The structure of EBOV L-VP35-RNA complex | | Descriptor: | Polymerase cofactor VP35, RNA-directed RNA polymerase L, The leader sequence of EBOV, ... | | Authors: | Qi, P, Yi, S. | | Deposit date: | 2023-06-20 | | Release date: | 2023-09-27 | | Last modified: | 2023-11-01 | | Method: | ELECTRON MICROSCOPY (2.95 Å) | | Cite: | Molecular mechanism of de novo replication by the Ebola virus polymerase.
Nature, 622, 2023
|
|
8JSM
 
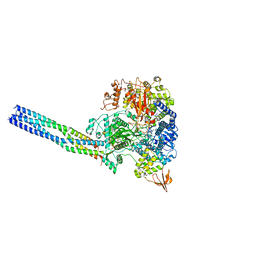 | | The structure of EBOV L-VP35-RNA complex (conformation 1) | | Descriptor: | Polymerase cofactor VP35, RNA-directed RNA polymerase L, The leader sequence of EBOV genome., ... | | Authors: | Qi, P, Yi, S. | | Deposit date: | 2023-06-20 | | Release date: | 2023-09-27 | | Last modified: | 2023-11-01 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Molecular mechanism of de novo replication by the Ebola virus polymerase.
Nature, 622, 2023
|
|
8JSN
 
 | | The structure of EBOV L-VP35-RNA complex (conformation 2) | | Descriptor: | Polymerase cofactor VP35, RNA-directed RNA polymerase L, The leader sequence of EBOV genome, ... | | Authors: | Qi, P, Yi, S. | | Deposit date: | 2023-06-20 | | Release date: | 2023-09-27 | | Last modified: | 2023-11-01 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Molecular mechanism of de novo replication by the Ebola virus polymerase.
Nature, 622, 2023
|
|
8IQC
 
 | |
8IQI
 
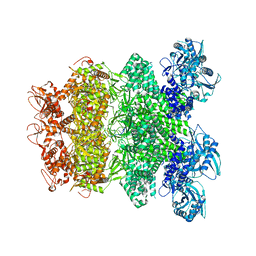 | | Structure of Full-Length AsfvPrimPol in Complex-Form | | Descriptor: | DNA (32-MER), MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ... | | Authors: | Shao, Z.W, Su, S.C, Gan, J.H. | | Deposit date: | 2023-03-16 | | Release date: | 2023-07-26 | | Last modified: | 2023-11-29 | | Method: | ELECTRON MICROSCOPY (3.32 Å) | | Cite: | Structures and implications of the C962R protein of African swine fever virus.
Nucleic Acids Res., 51, 2023
|
|
8IQH
 
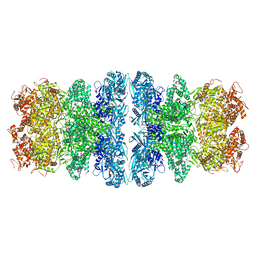 | |
8IQB
 
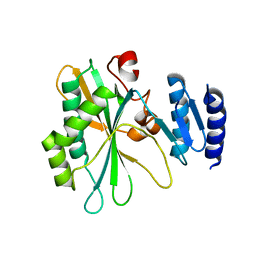 | |
8IQD
 
 | |
8JE2
 
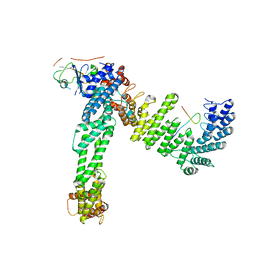 | | Cryo-EM structure of neddylated Cul2-Rbx1-EloBC-FEM1B complexed with FNIP1-FLCN | | Descriptor: | Cullin-2, Elongin-B, Elongin-C, ... | | Authors: | Dai, Z, Liang, L, Yin, Y.X. | | Deposit date: | 2023-05-15 | | Release date: | 2024-02-28 | | Last modified: | 2024-04-03 | | Method: | ELECTRON MICROSCOPY (3.63 Å) | | Cite: | Structural insights into the ubiquitylation strategy of the oligomeric CRL2 FEM1B E3 ubiquitin ligase.
Embo J., 43, 2024
|
|
8JE1
 
 | | An asymmetry dimer of the Cul2-Rbx1-EloBC-FEM1B ubiquitin ligase complexed with BEX2 | | Descriptor: | Cullin-2, E3 ubiquitin-protein ligase RBX1, Elongin-B, ... | | Authors: | Dai, Z, Liang, L, Yin, Y.X. | | Deposit date: | 2023-05-15 | | Release date: | 2024-02-28 | | Last modified: | 2024-04-03 | | Method: | ELECTRON MICROSCOPY (3.95 Å) | | Cite: | Structural insights into the ubiquitylation strategy of the oligomeric CRL2 FEM1B E3 ubiquitin ligase.
Embo J., 43, 2024
|
|
7XB0
 
 | | Crystal structure of Omicron BA.2 RBD complexed with hACE2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, ... | | Authors: | Li, L, Liao, H, Meng, Y, Li, W. | | Deposit date: | 2022-03-19 | | Release date: | 2022-07-06 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural basis of human ACE2 higher binding affinity to currently circulating Omicron SARS-CoV-2 sub-variants BA.2 and BA.1.1.
Cell, 185, 2022
|
|
