7OUI
 
 | | Structure of C2S2M2-type Photosystem supercomplex from Arabidopsis thaliana (digitonin-extracted) | | Descriptor: | (1R,3R)-6-{(3E,5E,7E,9E,11E,13E,15E,17E)-18-[(1S,4R,6R)-4-HYDROXY-2,2,6-TRIMETHYL-7-OXABICYCLO[4.1.0]HEPT-1-YL]-3,7,12,16-TETRAMETHYLOCTADECA-1,3,5,7,9,11,13,15,17-NONAENYLIDENE}-1,5,5-TRIMETHYLCYCLOHEXANE-1,3-DIOL, (3R,3'R,6S)-4,5-DIDEHYDRO-5,6-DIHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, (3S,5R,6S,3'S,5'R,6'S)-5,6,5',6'-DIEPOXY-5,6,5',6'- TETRAHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, ... | | Authors: | Graca, A.T, Hall, M, Persson, K, Schroder, W.P. | | Deposit date: | 2021-06-11 | | Release date: | 2021-08-18 | | Method: | ELECTRON MICROSCOPY (2.79 Å) | | Cite: | High-resolution model of Arabidopsis Photosystem II reveals the structural consequences of digitonin-extraction.
Sci Rep, 11, 2021
|
|
8OV8
 
 | | Crystal structure of Ene-reductase 1 from black poplar mushroom in complex to NADP | | Descriptor: | Ene-reductase 1, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, SODIUM ION, ... | | Authors: | Korf, L, Essen, L.-O, Karrer, D, Ruehl, M. | | Deposit date: | 2023-04-25 | | Release date: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Shifting the substrate scope of an ene/yne-reductase by loop engineering
To Be Published
|
|
8F43
 
 | | HNH Nuclease Domain from G. stearothermophilus Cas9, K597A mutant | | Descriptor: | CRISPR-associated endonuclease Cas9 | | Authors: | D'Ordine, A.M, Belato, H.B, Lisi, G.P, Jogl, G. | | Deposit date: | 2022-11-10 | | Release date: | 2022-12-21 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.37 Å) | | Cite: | Disruption of electrostatic contacts in the HNH nuclease from a thermophilic Cas9 rewires allosteric motions and enhances high-temperature DNA cleavage.
J.Chem.Phys., 157, 2022
|
|
8PHV
 
 | | Structure of Human selenomethionylated Cdc123 bound to domain 3 of eIF2 gamma | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Cell division cycle protein 123 homolog, Eukaryotic translation initiation factor 2 subunit 3 | | Authors: | Schmitt, E, Mechulam, Y, Cardenal Peralta, C, Fagart, J, Seufert, W. | | Deposit date: | 2023-06-20 | | Release date: | 2023-08-16 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Binding of human Cdc123 to eIF2 gamma.
J.Struct.Biol., 215, 2023
|
|
7OS2
 
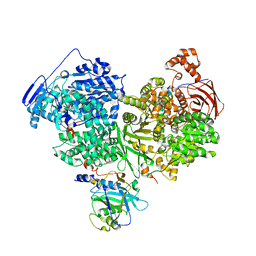 | | Cryo-EM structure of Brr2 in complex with Jab1/MPN and C9ORF78 | | Descriptor: | Pre-mRNA-processing-splicing factor 8, Telomere length and silencing protein 1 homolog, U5 small nuclear ribonucleoprotein 200 kDa helicase | | Authors: | Bergfort, A, Hilal, T, Weber, G, Wahl, M.C. | | Deposit date: | 2021-06-07 | | Release date: | 2022-02-23 | | Last modified: | 2024-09-25 | | Method: | ELECTRON MICROSCOPY (2.76 Å) | | Cite: | The intrinsically disordered TSSC4 protein acts as a helicase inhibitor, placeholder and multi-interaction coordinator during snRNP assembly and recycling.
Nucleic Acids Res., 50, 2022
|
|
7OS1
 
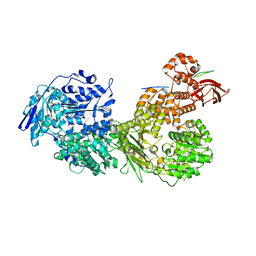 | | Cryo-EM structure of Brr2 in complex with Fbp21 | | Descriptor: | U5 small nuclear ribonucleoprotein 200 kDa helicase, WW domain-binding protein 4 | | Authors: | Bergfort, A, Hilal, T, Weber, G, Wahl, M.C. | | Deposit date: | 2021-06-07 | | Release date: | 2022-02-23 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | The intrinsically disordered TSSC4 protein acts as a helicase inhibitor, placeholder and multi-interaction coordinator during snRNP assembly and recycling.
Nucleic Acids Res., 50, 2022
|
|
8PKU
 
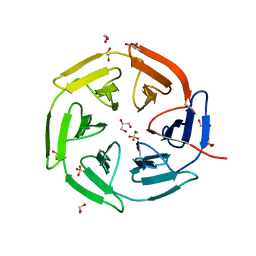 | | Kelch domain of KEAP1 in complex with ortho-dimethylbenzene linked cyclic peptide 3 (ortho-WRCDEETGEC). | | Descriptor: | (2-methylphenyl)methanol, 1,2-ETHANEDIOL, CHLORIDE ION, ... | | Authors: | Braun, M.B, Bischof, L, Hartmann, M.D. | | Deposit date: | 2023-06-27 | | Release date: | 2023-11-15 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Computational Prediction of Cyclic Peptide Structural Ensembles and Application to the Design of Keap1 Binders.
J.Chem.Inf.Model., 63, 2023
|
|
8F6U
 
 | |
8F6V
 
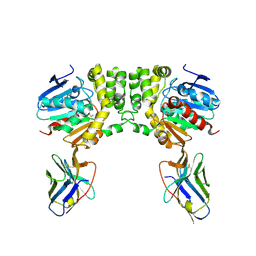 | |
8PKC
 
 | |
8PVZ
 
 | |
7K40
 
 | | Crystal Structure of SARS-CoV-2 Main Protease (3CLpro/Mpro) in Complex with Covalent Inhibitor Boceprevir at 1.35 A Resolution | | Descriptor: | 3C-like proteinase, DIMETHYL SULFOXIDE, boceprevir (bound form) | | Authors: | Kumaran, D, Andi, B, Kreitler, D.F, Soares, A.S, Shi, W, Jakoncic, J, Fuchs, M.R, Keereetaweep, J, Shanklin, J, McSweeney, S. | | Deposit date: | 2020-09-14 | | Release date: | 2020-09-23 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Hepatitis C virus NS3/4A inhibitors and other drug-like compounds as covalent binders of SARS-CoV-2 main protease.
Sci Rep, 12, 2022
|
|
8GAR
 
 | |
7JYC
 
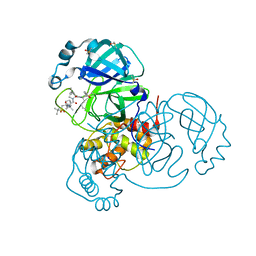 | | Crystal Structure of SARS-CoV-2 Main Protease (3CLpro/Mpro) in Complex with Covalent Inhibitor Narlaprevir | | Descriptor: | (1R,2S,5S)-3-[N-({1-[(tert-butylsulfonyl)methyl]cyclohexyl}carbamoyl)-3-methyl-L-valyl]-N-{(1S)-1-[(1R)-2-(cyclopropylamino)-1-hydroxy-2-oxoethyl]pentyl}-6,6-dimethyl-3-azabicyclo[3.1.0]hexane-2-carboxamide, 3C-like proteinase, DIMETHYL SULFOXIDE, ... | | Authors: | Andi, B, Kumaran, D, Kreitler, D.F, Soares, A.S, Shi, W, Jakoncic, J, Fuchs, M.R, Keereetaweep, J, Shanklin, J, McSweeney, S. | | Deposit date: | 2020-08-30 | | Release date: | 2020-09-09 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Hepatitis C virus NS3/4A inhibitors and other drug-like compounds as covalent binders of SARS-CoV-2 main protease.
Sci Rep, 12, 2022
|
|
8Q6B
 
 | | The RSL-D32N - sulfonato-calix[8]arene complex, I23 form, citrate pH 4.0, obtained by cross-seeding | | Descriptor: | Fucose-binding lectin protein, GLYCEROL, beta-D-fructopyranose, ... | | Authors: | Flood, R.J, Crowley, P.B. | | Deposit date: | 2023-08-11 | | Release date: | 2024-03-06 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | Supramolecular Synthons in Protein-Ligand Frameworks.
Cryst.Growth Des., 24, 2024
|
|
8GJR
 
 | |
7K6D
 
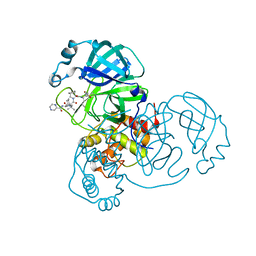 | | SARS-CoV-2 Main Protease Co-Crystal Structure with Telaprevir Determined from Crystals Grown with 40 nL Acoustically Ejected Mpro Droplets at 1.48 A Resolution (Cryo-protected) | | Descriptor: | (1S,3aR,6aS)-2-[(2S)-2-({(2S)-2-cyclohexyl-2-[(pyrazin-2-ylcarbonyl)amino]acetyl}amino)-3,3-dimethylbutanoyl]-N-[(2R,3S)-1-(cyclopropylamino)-2-hydroxy-1-oxohexan-3-yl]octahydrocyclopenta[c]pyrrole-1-carboxamide, 3C-like proteinase, DIMETHYL SULFOXIDE | | Authors: | Kreitler, D.F, Andi, B, Kumaran, D, Soares, A.S, Shi, W, Jakoncic, J, Fuchs, M.R, Keereetaweep, J, Shanklin, J, McSweeney, S. | | Deposit date: | 2020-09-19 | | Release date: | 2020-09-30 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Hepatitis C virus NS3/4A inhibitors and other drug-like compounds as covalent binders of SARS-CoV-2 main protease.
Sci Rep, 12, 2022
|
|
6WYG
 
 | |
6Y87
 
 | |
7K3T
 
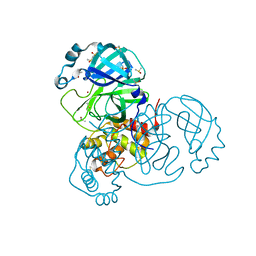 | | Crystal Structure of SARS-CoV-2 Main Protease (3CLpro/Mpro) at 1.2 A Resolution and a Possible Capture of Zinc Binding Intermediate | | Descriptor: | 3C-like proteinase, DIMETHYL SULFOXIDE, ZINC ION | | Authors: | Andi, B, Kumaran, D, Kreitler, D.F, Soares, A.S, Shi, W, Jakoncic, J, Fuchs, M.R, Keereetaweep, J, Shanklin, J, McSweeney, S. | | Deposit date: | 2020-09-13 | | Release date: | 2020-09-23 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Hepatitis C virus NS3/4A inhibitors and other drug-like compounds as covalent binders of SARS-CoV-2 main protease.
Sci Rep, 12, 2022
|
|
7PTH
 
 | |
7PJH
 
 | | Crystal structure of the human spliceosomal maturation factor AAR2 bound to the RNAse H domain of PRPF8 | | Descriptor: | Pre-mRNA-processing-splicing factor 8, Protein AAR2 homolog | | Authors: | Preussner, M, Santos, K, Heroven, A.C, Alles, J, Heyd, F, Wahl, M.C, Weber, G. | | Deposit date: | 2021-08-24 | | Release date: | 2022-11-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structural and functional investigation of the human snRNP assembly factor AAR2 in complex with the RNase H-like domain of PRPF8.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
8SXS
 
 | |
8SCA
 
 | | Rec3 Domain from S. pyogenes Cas9 | | Descriptor: | 1,2-ETHANEDIOL, CRISPR-associated endonuclease Cas9/Csn1 | | Authors: | D'Ordine, A.M, Skeens, E, Lisi, G.P, Jogl, G. | | Deposit date: | 2023-04-05 | | Release date: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | High-fidelity, hyper-accurate, and evolved mutants rewire atomic-level communication in CRISPR-Cas9.
Sci Adv, 10, 2024
|
|
7LM4
 
 | | The crystal structure of the I38T mutant PA Endonuclease (2009/H1N1/CALIFORNIA) in complex with SJ000988503 | | Descriptor: | 5-hydroxy-N-[2-(4-hydroxy-3-methoxyphenyl)ethyl]-2-(2-methylphenyl)-6-oxo-1,6-dihydropyrimidine-4-carboxamide, Hexa Vinylpyrrolidone K15, MANGANESE (II) ION, ... | | Authors: | Cuypers, M.G, Slavish, P.J, Jayaraman, S, Rankovic, Z, White, S.W. | | Deposit date: | 2021-02-05 | | Release date: | 2022-02-09 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Chemical scaffold recycling: Structure-guided conversion of an HIV integrase inhibitor into a potent influenza virus RNA-dependent RNA polymerase inhibitor designed to minimize resistance potential.
Eur.J.Med.Chem., 247, 2023
|
|
