6P8J
 
 | |
6P8U
 
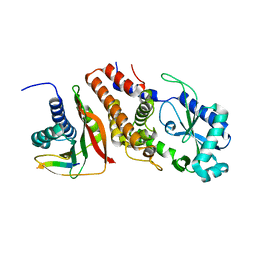 | | Structure of P. aeruginosa ATCC27853 CdnD:HORMA2:Peptide 1 complex | | Descriptor: | HORMA domain containing protein, MAGNESIUM ION, Nucleotidyltransferase, ... | | Authors: | Ye, Q, Corbett, K.D. | | Deposit date: | 2019-06-08 | | Release date: | 2019-12-25 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.893 Å) | | Cite: | HORMA Domain Proteins and a Trip13-like ATPase Regulate Bacterial cGAS-like Enzymes to Mediate Bacteriophage Immunity.
Mol.Cell, 77, 2020
|
|
6P8S
 
 | |
4XHH
 
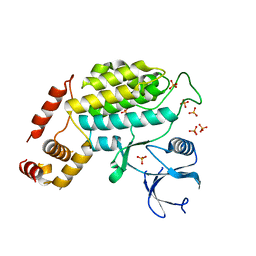 | | Structure of C. glabrata Hrr25, Apo state | | Descriptor: | PHOSPHATE ION, Similar to uniprot|P29295 Saccharomyces cerevisiae YPL204w HRR25 | | Authors: | Ye, Q, Corbett, K.D. | | Deposit date: | 2015-01-05 | | Release date: | 2016-01-06 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.908 Å) | | Cite: | Structure of the Saccharomyces cerevisiae Hrr25:Mam1 monopolin subcomplex reveals a novel kinase regulator.
Embo J., 35, 2016
|
|
4XHL
 
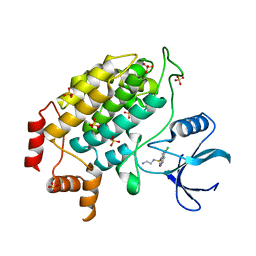 | | Structure of S. cerevisiae Hrr25 1-394 (K38R mutant) | | Descriptor: | Casein kinase I homolog HRR25, N-(2-AMINOETHYL)-5-CHLOROISOQUINOLINE-8-SULFONAMIDE, SULFATE ION | | Authors: | Ye, Q, Corbett, K.D. | | Deposit date: | 2015-01-05 | | Release date: | 2016-01-06 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.01 Å) | | Cite: | Structure of the Saccharomyces cerevisiae Hrr25:Mam1 monopolin subcomplex reveals a novel kinase regulator.
Embo J., 35, 2016
|
|
4XGU
 
 | | Structure of C. elegans PCH-2 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Putative pachytene checkpoint protein 2, SULFATE ION | | Authors: | Ye, Q, Corbett, K.D. | | Deposit date: | 2015-01-02 | | Release date: | 2015-05-06 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.301 Å) | | Cite: | TRIP13 is a protein-remodeling AAA+ ATPase that catalyzes MAD2 conformation switching.
Elife, 4, 2015
|
|
4XH0
 
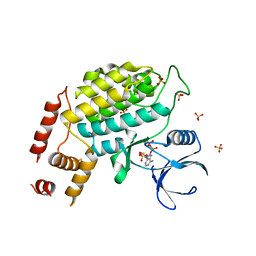 | | Structure of C. glabrata Hrr25 bound to ADP (SO4 condition) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, SULFATE ION, Similar to uniprot|P29295 Saccharomyces cerevisiae YPL204w HRR25 | | Authors: | Ye, Q, Corbett, K.D. | | Deposit date: | 2015-01-04 | | Release date: | 2016-01-06 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Structure of the Saccharomyces cerevisiae Hrr25:Mam1 monopolin subcomplex reveals a novel kinase regulator.
Embo J., 35, 2016
|
|
4XHG
 
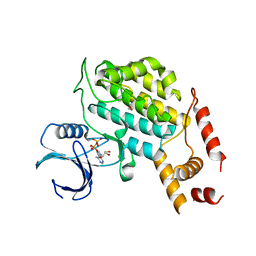 | | Structure of C. glabrata Hrr25 bound to ADP (formate condition) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, FORMIC ACID, Similar to uniprot|P29295 Saccharomyces cerevisiae YPL204w HRR25 | | Authors: | Ye, Q, Corbett, K.D. | | Deposit date: | 2015-01-05 | | Release date: | 2016-01-06 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structure of the Saccharomyces cerevisiae Hrr25:Mam1 monopolin subcomplex reveals a novel kinase regulator.
Embo J., 35, 2016
|
|
1C9W
 
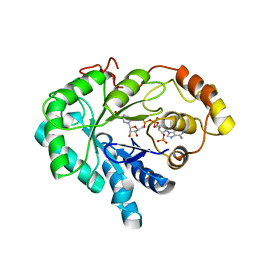 | | CHO REDUCTASE WITH NADP+ | | Descriptor: | CHO REDUCTASE, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Ye, Q, Li, X, Hyndman, D, Flynn, T.G, Jia, Z. | | Deposit date: | 1999-08-03 | | Release date: | 2000-01-07 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of CHO reductase, a member of the aldo-keto reductase superfamily.
Proteins, 38, 2000
|
|
5E9E
 
 | |
5VQ9
 
 | | Structure of human TRIP13, Apo form | | Descriptor: | Pachytene checkpoint protein 2 homolog | | Authors: | Ye, Q, Corbett, K.D. | | Deposit date: | 2017-05-08 | | Release date: | 2017-06-14 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.02 Å) | | Cite: | The AAA+ ATPase TRIP13 remodels HORMA domains through N-terminal engagement and unfolding.
EMBO J., 36, 2017
|
|
5CYZ
 
 | | Structure of S. cerevisiae Hrr25:Mam1 complex, form 1 | | Descriptor: | Casein kinase I homolog HRR25, Monopolin complex subunit MAM1, ZINC ION | | Authors: | Ye, Q, Corbett, K.D. | | Deposit date: | 2015-07-31 | | Release date: | 2016-08-03 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.841 Å) | | Cite: | Structure of the Saccharomyces cerevisiae Hrr25:Mam1 monopolin subcomplex reveals a novel kinase regulator.
Embo J., 35, 2016
|
|
5E4H
 
 | |
5VQA
 
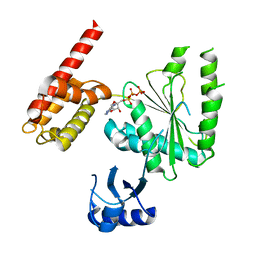 | | Structure of human TRIP13, ATP-bound form | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Pachytene checkpoint protein 2 homolog | | Authors: | Ye, Q, Corbett, K.D. | | Deposit date: | 2017-05-08 | | Release date: | 2017-06-14 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.54 Å) | | Cite: | The AAA+ ATPase TRIP13 remodels HORMA domains through N-terminal engagement and unfolding.
EMBO J., 36, 2017
|
|
5CZO
 
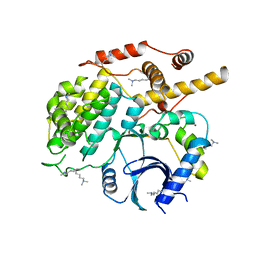 | | Structure of S. cerevisiae Hrr25:Mam1 complex, form 2 | | Descriptor: | Casein kinase I homolog HRR25, Monopolin complex subunit MAM1, ZINC ION | | Authors: | Ye, Q, Corbett, K.D. | | Deposit date: | 2015-07-31 | | Release date: | 2016-08-03 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.894 Å) | | Cite: | Structure of the Saccharomyces cerevisiae Hrr25:Mam1 monopolin subcomplex reveals a novel kinase regulator.
Embo J., 35, 2016
|
|
4IRY
 
 | |
6BDT
 
 | |
6BGP
 
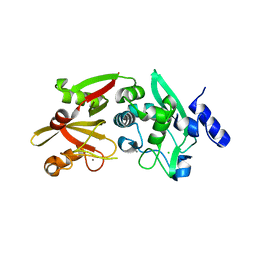 | |
6BJD
 
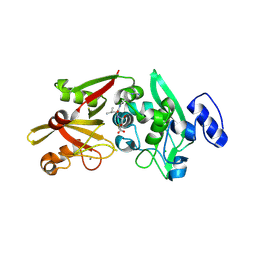 | | Crystal Structure of Human Calpain-3 Protease Core in Complex with E-64 | | Descriptor: | CALCIUM ION, CHLORIDE ION, Calpain-3, ... | | Authors: | Ye, Q, Campbell, R.L, Davies, P.L. | | Deposit date: | 2017-11-06 | | Release date: | 2018-02-07 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structures of human calpain-3 protease core with and without bound inhibitor reveal mechanisms of calpain activation.
J. Biol. Chem., 293, 2018
|
|
6BKJ
 
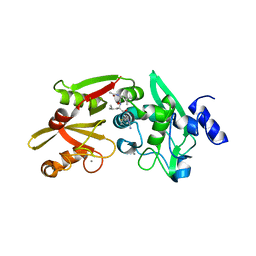 | |
7R98
 
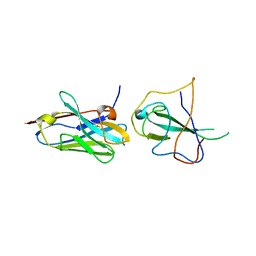 | |
6UXF
 
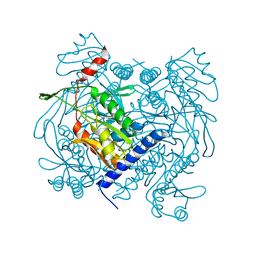 | | Structure of V. metoecus NucC, hexamer form | | Descriptor: | Vibrio meotecus sp. RC341 NucC | | Authors: | Ye, Q, Corbett, K.D. | | Deposit date: | 2019-11-07 | | Release date: | 2019-12-25 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structure and Mechanism of a Cyclic Trinucleotide-Activated Bacterial Endonuclease Mediating Bacteriophage Immunity.
Mol.Cell, 77, 2020
|
|
6UXG
 
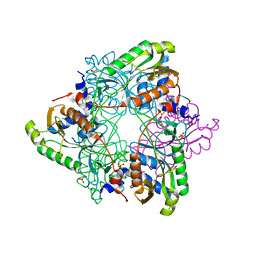 | | Structure of V. metoecus NucC, trimer form | | Descriptor: | SULFATE ION, Vibrio metoecus sp. RC341 NucC | | Authors: | Ye, Q, Corbett, K.D. | | Deposit date: | 2019-11-07 | | Release date: | 2019-12-25 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structure and Mechanism of a Cyclic Trinucleotide-Activated Bacterial Endonuclease Mediating Bacteriophage Immunity.
Mol.Cell, 77, 2020
|
|
6XI3
 
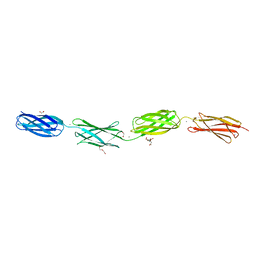 | | Crystal structure of tetra-tandem repeat in extending region of large adhesion protein | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Ye, Q, Vance, T.D.R, Davies, P.L. | | Deposit date: | 2020-06-19 | | Release date: | 2020-10-14 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Essential role of calcium in extending RTX adhesins to their target.
J Struct Biol X, 4, 2020
|
|
6XI1
 
 | | Crystal structure of tetra-tandem repeat in extending RTX adhesin from Aeromonas hydrophila | | Descriptor: | CALCIUM ION, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Ye, Q, Vance, T.D.R, Conroy, B, Davies, P.L. | | Deposit date: | 2020-06-19 | | Release date: | 2020-10-14 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Essential role of calcium in extending RTX adhesins to their target.
J Struct Biol X, 4, 2020
|
|
