3QC8
 
 | | Crystal Structure of FAF1 UBX Domain In Complex with p97/VCP N Domain Reveals The Conserved FcisP Touch-Turn Motif of UBX Domain Suffering Conformational Change | | Descriptor: | FAS-associated factor 1, Transitional endoplasmic reticulum ATPase | | Authors: | Kim, K.H, Kang, W, Suh, S.W, Yang, J.K. | | Deposit date: | 2011-01-15 | | Release date: | 2011-07-20 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of FAF1 UBX domain in complex with p97/VCP N domain reveals a conformational change in the conserved FcisP touch-turn motif of UBX domain
Proteins, 79, 2011
|
|
3QCA
 
 | |
3NX2
 
 | | Enterobacter sp. Px6-4 Ferulic Acid Decarboxylase in complex with substrate analogues | | Descriptor: | 3-(4-HYDROXY-3-METHOXYPHENYL)-2-PROPENOIC ACID, Ferulic acid decarboxylase | | Authors: | Gu, W, Yang, J.K, Lou, Z.Y, Meng, Z.H, Zhang, K.-Q. | | Deposit date: | 2010-07-12 | | Release date: | 2011-02-16 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Structural Basis of Enzymatic Activity for the Ferulic Acid Decarboxylase (FADase) from Enterobacter sp. Px6-4
Plos One, 6, 2011
|
|
3NX1
 
 | | Crystal structure of Enterobacter sp. Px6-4 Ferulic Acid Decarboxylase | | Descriptor: | Ferulic acid decarboxylase | | Authors: | Gu, W, Yang, J.K, Lou, Z.Y, Meng, Z.H, Zhang, K.-Q. | | Deposit date: | 2010-07-12 | | Release date: | 2011-02-16 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural Basis of Enzymatic Activity for the Ferulic Acid Decarboxylase (FADase) from Enterobacter sp. Px6-4
Plos One, 6, 2011
|
|
4M6R
 
 | | Structural and biochemical basis for the inhibition of cell death by APIP, a methionine salvage enzyme | | Descriptor: | Methylthioribulose-1-phosphate dehydratase, ZINC ION | | Authors: | Kang, W, Hong, S.H, Lee, H.M, Kim, N.Y, Lim, Y.C, Le, L.T.M, Lim, B, Kim, H.C, Kim, T.Y, Ashida, H, Yokota, A, Hah, S.S, Chun, K.H, Jung, Y.K, Yang, J.K. | | Deposit date: | 2013-08-10 | | Release date: | 2014-01-29 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural and biochemical basis for the inhibition of cell death by APIP, a methionine salvage enzyme.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
1DGS
 
 | | CRYSTAL STRUCTURE OF NAD+-DEPENDENT DNA LIGASE FROM T. FILIFORMIS | | Descriptor: | ADENOSINE MONOPHOSPHATE, DNA LIGASE, ZINC ION | | Authors: | Lee, J.Y, Chang, C, Song, H.K, Kwon, S.T, Suh, S.W. | | Deposit date: | 1999-11-25 | | Release date: | 2000-11-27 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal structure of NAD(+)-dependent DNA ligase: modular architecture and functional implications.
EMBO J., 19, 2000
|
|
1C02
 
 | | CRYSTAL STRUCTURE OF YEAST YPD1P | | Descriptor: | PHOSPHOTRANSFERASE YPD1P | | Authors: | Song, H.K, Lee, J.Y, Lee, M.G, Suh, S.W. | | Deposit date: | 1999-07-14 | | Release date: | 2000-01-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Insights into eukaryotic multistep phosphorelay signal transduction revealed by the crystal structure of Ypd1p from Saccharomyces cerevisiae.
J.Mol.Biol., 293, 1999
|
|
1C03
 
 | | CRYSTAL STRUCTURE OF YPD1P (TRICLINIC FORM) | | Descriptor: | HYPOTHETICAL PROTEIN YDL235C | | Authors: | Song, H.K, Lee, J.Y, Lee, M.G, Suh, S.W. | | Deposit date: | 1999-07-14 | | Release date: | 2000-01-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Insights into eukaryotic multistep phosphorelay signal transduction revealed by the crystal structure of Ypd1p from Saccharomyces cerevisiae.
J.Mol.Biol., 293, 1999
|
|
5F8E
 
 | | Rv2258c-SAH | | Descriptor: | Methyltransferase, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Im, H.N, Suh, S.W. | | Deposit date: | 2015-12-09 | | Release date: | 2016-06-29 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal structure of Rv2258c from Mycobacterium tuberculosis H37Rv, an S-adenosyl-l-methionine-dependent methyltransferase
J.Struct.Biol., 193, 2016
|
|
5F8C
 
 | | Rv2258c-unbound | | Descriptor: | GLYCEROL, Methyltransferase | | Authors: | Im, H.N, Suh, S.W. | | Deposit date: | 2015-12-09 | | Release date: | 2016-06-29 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Crystal structure of Rv2258c from Mycobacterium tuberculosis H37Rv, an S-adenosyl-l-methionine-dependent methyltransferase
J.Struct.Biol., 193, 2016
|
|
5F8F
 
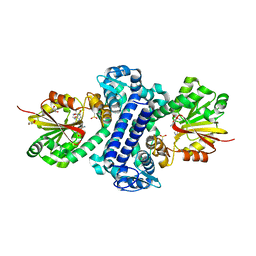 | | Rv2258c-SFG | | Descriptor: | GLYCEROL, Methyltransferase, SINEFUNGIN | | Authors: | Im, H.N, Suh, S.W. | | Deposit date: | 2015-12-09 | | Release date: | 2016-06-29 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of Rv2258c from Mycobacterium tuberculosis H37Rv, an S-adenosyl-l-methionine-dependent methyltransferase
J.Struct.Biol., 193, 2016
|
|
7WGZ
 
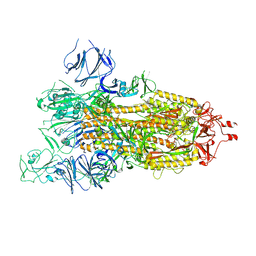 | | SARS-CoV-2 spike glycoprotein trimer in open state | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Zhu, Y, Tai, L, Yin, G, Sun, F. | | Deposit date: | 2021-12-29 | | Release date: | 2023-01-04 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | Novel cleavage sites identified in SARS-CoV-2 spike protein reveal mechanism for cathepsin L-facilitated viral infection and treatment strategies
Cell Discov, 8, 2022
|
|
7WGY
 
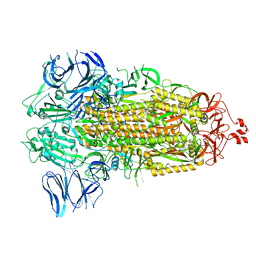 | | SARS-CoV-2 spike glycoprotein trimer in Intermediate state | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Zhu, Y, Tai, L, Yin, G, Sun, F. | | Deposit date: | 2021-12-29 | | Release date: | 2023-01-04 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Novel cleavage sites identified in SARS-CoV-2 spike protein reveal mechanism for cathepsin L-facilitated viral infection and treatment strategies
Cell Discov, 8, 2022
|
|
7WGV
 
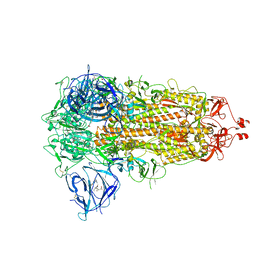 | | SARS-CoV-2 spike glycoprotein trimer in closed state | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 3-[5-[(Z)-(4-ethenyl-3-methyl-5-oxidanylidene-pyrrol-2-ylidene)methyl]-2-[[5-[(Z)-(3-ethenyl-4-methyl-5-oxidanylidene-pyrrol-2-ylidene)methyl]-3-(3-hydroxy-3-oxopropyl)-4-methyl-1H-pyrrol-2-yl]methyl]-4-methyl-1H-pyrrol-3-yl]propanoic acid, ... | | Authors: | Zhu, Y, Tai, L, Yin, G, Sun, F. | | Deposit date: | 2021-12-29 | | Release date: | 2023-01-04 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Novel cleavage sites identified in SARS-CoV-2 spike protein reveal mechanism for cathepsin L-facilitated viral infection and treatment strategies
Cell Discov, 8, 2022
|
|
7WGX
 
 | | SARS-CoV-2 spike glycoprotein trimer in closed state after treatment with Cathepsin L | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 3-[5-[(Z)-(4-ethenyl-3-methyl-5-oxidanylidene-pyrrol-2-ylidene)methyl]-2-[[5-[(Z)-(3-ethenyl-4-methyl-5-oxidanylidene-pyrrol-2-ylidene)methyl]-3-(3-hydroxy-3-oxopropyl)-4-methyl-1H-pyrrol-2-yl]methyl]-4-methyl-1H-pyrrol-3-yl]propanoic acid, ... | | Authors: | Zhu, Y, Tai, L, Yin, G, Sun, F. | | Deposit date: | 2021-12-29 | | Release date: | 2023-01-04 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Novel cleavage sites identified in SARS-CoV-2 spike protein reveal mechanism for cathepsin L-facilitated viral infection and treatment strategies
Cell Discov, 8, 2022
|
|
2EFF
 
 | |
3OQ9
 
 | | Structure of the FAS/FADD death domain assembly | | Descriptor: | Protein FADD, Tumor necrosis factor receptor superfamily member 6 | | Authors: | Kabaleeswaran, V, Wu, H. | | Deposit date: | 2010-09-02 | | Release date: | 2010-10-13 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (6.8 Å) | | Cite: | The Fas-FADD death domain complex structure reveals the basis of DISC assembly and disease mutations.
Nat.Struct.Mol.Biol., 17, 2010
|
|
1JXV
 
 | | Crystal Structure of Human Nucleoside Diphosphate Kinase A | | Descriptor: | Nucleoside Diphosphate Kinase A | | Authors: | Min, K, Song, H.K, Chang, C, Kim, S.Y, Lee, K.J, Suh, S.W. | | Deposit date: | 2001-09-10 | | Release date: | 2002-04-10 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of human nucleoside diphosphate kinase A, a metastasis suppressor.
Proteins, 46, 2002
|
|
1TAE
 
 | |
1IX1
 
 | | Crystal Structure of P.aeruginosa Peptide deformylase Complexed with Antibiotic Actinonin | | Descriptor: | (CARBAMOYLMETHYL-CARBOXYMETHYL-AMINO)-ACETIC ACID, ACTINONIN, ZINC ION, ... | | Authors: | Kim, H.-W, Yoon, H.-J, Lee, J.Y, Han, B.W, Yang, J.K, Lee, B.I, Ahn, H.J, Lee, H.H, Suh, S.W. | | Deposit date: | 2002-06-07 | | Release date: | 2003-09-09 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal structure of peptide deformylase from Staphylococcus aureus in complex with actinonin, a naturally occurring antibacterial agent
Proteins, 57, 2004
|
|
1TA8
 
 | |
1TX6
 
 | | trypsin:BBI complex | | Descriptor: | Bowman-Birk type trypsin inhibitor, CALCIUM ION, Trypsin | | Authors: | Song, H.K, Park, E.Y, Kim, J.A, Kim, H.W, Kim, Y.S. | | Deposit date: | 2004-07-02 | | Release date: | 2005-03-08 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of the Bowman-Birk inhibitor from barley seeds in ternary complex with porcine trypsin
J.Mol.Biol., 343, 2004
|
|
2P1X
 
 | |
