2XWS
 
 | | ANAEROBIC COBALT CHELATASE (CbiX) FROM ARCHAEOGLOBUS FULGIDUS | | Descriptor: | SIROHYDROCHLORIN COBALTOCHELATASE | | Authors: | Romao, C.V, Ladakis, D, Lobo, S.A.L, Carrondo, M.A, Brindley, A.A, Deery, E, Matias, P.M, Pickersgill, R.W, Saraiva, L.M, Warren, M.J. | | Deposit date: | 2010-11-04 | | Release date: | 2010-12-22 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Evolution in a Family of Chelatases Facilitated by the Introduction of Active Site Asymmetry and Protein Oligomerization.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
1QGO
 
 | | ANAEROBIC COBALT CHELATASE IN COBALAMIN BIOSYNTHESIS FROM SALMONELLA TYPHIMURIUM | | Descriptor: | ANAEROBIC COBALAMIN BIOSYNTHETIC COBALT CHELATASE, SULFATE ION | | Authors: | Schubert, H.L, Raux, E, Warren, M.J, Wilson, K.S. | | Deposit date: | 1999-05-03 | | Release date: | 1999-09-29 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Common chelatase design in the branched tetrapyrrole pathways of heme and anaerobic cobalamin synthesis.
Biochemistry, 38, 1999
|
|
2CBF
 
 | | THE X-RAY STRUCTURE OF A COBALAMIN BIOSYNTHETIC ENZYME, COBALT PRECORRIN-4 METHYLTRANSFERASE, CBIF, FROM BACILLUS MEGATERIUM, WITH THE HIS-TAG CLEAVED OFF | | Descriptor: | COBALT-PRECORRIN-4 TRANSMETHYLASE, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Schubert, H.L, Raux, E, Woodcock, S.C, Warren, M.J, Wilson, K.S. | | Deposit date: | 1998-05-01 | | Release date: | 1999-05-11 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | The X-ray structure of a cobalamin biosynthetic enzyme, cobalt-precorrin-4 methyltransferase.
Nat.Struct.Biol., 5, 1998
|
|
1CBF
 
 | | THE X-RAY STRUCTURE OF A COBALAMIN BIOSYNTHETIC ENZYME, COBALT PRECORRIN-4 METHYLTRANSFERASE, CBIF | | Descriptor: | COBALT-PRECORRIN-4 TRANSMETHYLASE, PHOSPHATE ION, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Schubert, H.L, Raux, E, Woodcock, S.C, Wilson, K.S, Warren, M.J. | | Deposit date: | 1998-05-01 | | Release date: | 1999-05-11 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The X-ray structure of a cobalamin biosynthetic enzyme, cobalt-precorrin-4 methyltransferase.
Nat.Struct.Biol., 5, 1998
|
|
2QBU
 
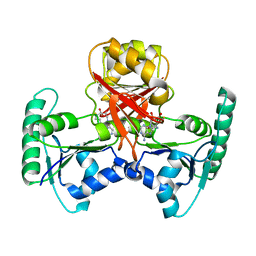 | | Crystal structure of Methanothermobacter thermautotrophicus CbiL | | Descriptor: | Precorrin-2 methyltransferase, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Frank, S, Warren, M.J, Pickersgill, R.W. | | Deposit date: | 2007-06-18 | | Release date: | 2008-04-15 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Elucidation of substrate specificity in the cobalamin (vitamin B12) biosynthetic methyltransferases. Structure and function of the C20 methyltransferase (CbiL) from Methanothermobacter thermautotrophicus.
J.Biol.Chem., 282, 2007
|
|
4CCS
 
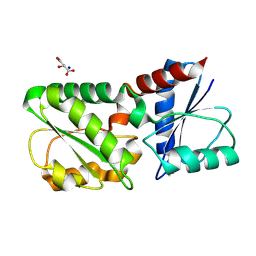 | | The structure of CbiX, the terminal Enzyme for Biosynthesis of Siroheme in Denitrifying Bacteria | | Descriptor: | 1,2-ETHANEDIOL, CBIX, D-MALATE, ... | | Authors: | Bali, S, Rollauer, S.E, Roversi, P, Raux-Deery, E, Lea, S.M, Warren, M.J, Ferguson, S.J. | | Deposit date: | 2013-10-25 | | Release date: | 2014-04-09 | | Last modified: | 2015-09-23 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Identification and Characterization of the 'Missing' Terminal Enzyme for Siroheme Biosynthesis in Alpha-Proteobacteria.
Mol.Microbiol., 92, 2014
|
|
4CZD
 
 | |
2W6L
 
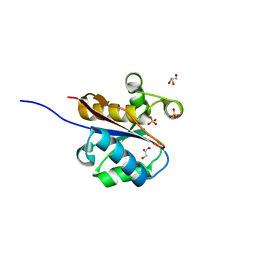 | | The crystal structure at 1.7 A resolution of CobE, a protein from the cobalamin (vitamin B12) biosynthetic pathway | | Descriptor: | COBE, GLYCEROL, SULFATE ION | | Authors: | Vevodova, J, Smith, D, McGoldrick, H, Deery, E, Murzin, A.G, Warren, M.J, Wilson, K.S. | | Deposit date: | 2008-12-18 | | Release date: | 2008-12-30 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | The Crystal Structure at 1.7 A Resolution of Cobe, a Protein from the Cobalamin (Vitamin B12) Biosynthetic Pathway
To be Published
|
|
1PDA
 
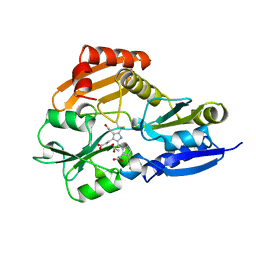 | | STRUCTURE OF PORPHOBILINOGEN DEAMINASE REVEALS A FLEXIBLE MULTIDOMAIN POLYMERASE WITH A SINGLE CATALYTIC SITE | | Descriptor: | 3-[5-{[3-(2-carboxyethyl)-4-(carboxymethyl)-5-methyl-1H-pyrrol-2-yl]methyl}-4-(carboxymethyl)-1H-pyrrol-3-yl]propanoic acid, ACETIC ACID, PORPHOBILINOGEN DEAMINASE | | Authors: | Louie, G.V, Brownlie, P.D, Lambert, R, Cooper, J.B, Blundell, T.L, Wood, S.P, Warren, M.J, Woodcock, S.C, Jordan, P.M. | | Deposit date: | 1992-11-17 | | Release date: | 1993-10-31 | | Last modified: | 2019-08-14 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Structure of porphobilinogen deaminase reveals a flexible multidomain polymerase with a single catalytic site.
Nature, 359, 1992
|
|
1PJS
 
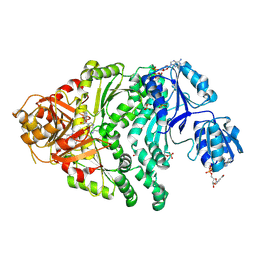 | | The co-crystal structure of CysG, the multifunctional methyltransferase/dehydrogenase/ferrochelatase for siroheme synthesis, in complex with it NAD cofactor | | Descriptor: | NICOTINAMIDE-ADENINE-DINUCLEOTIDE, PHOSPHATE ION, S-ADENOSYL-L-HOMOCYSTEINE, ... | | Authors: | Stroupe, M.E, Leech, H.K, Daniels, D.S, Warren, M.J, Getzoff, E.D. | | Deposit date: | 2003-06-03 | | Release date: | 2003-12-02 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | CysG structure reveals tetrapyrrole-binding features and novel regulation of siroheme biosynthesis.
Nat.Struct.Biol., 10, 2003
|
|
1PJQ
 
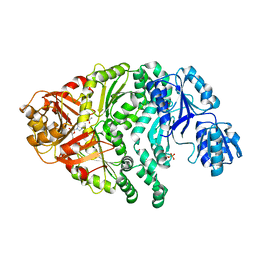 | | Structure and function of CysG, the multifunctional methyltransferase/dehydrogenase/ferrochelatase for siroheme synthesis | | Descriptor: | ACETATE ION, S-ADENOSYL-L-HOMOCYSTEINE, Siroheme synthase, ... | | Authors: | Stroupe, M.E, Leech, H.K, Daniels, D.S, Warren, M.J, Getzoff, E.D. | | Deposit date: | 2003-06-03 | | Release date: | 2003-12-02 | | Last modified: | 2014-11-12 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | CysG structure reveals tetrapyrrole-binding features and novel regulation of siroheme biosynthesis.
Nat.Struct.Biol., 10, 2003
|
|
1PJT
 
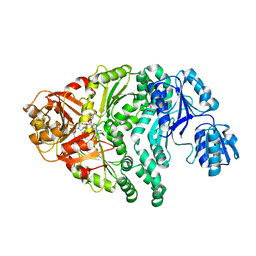 | | The structure of the Ser128Ala point-mutant variant of CysG, the multifunctional methyltransferase/dehydrogenase/ferrochelatase for siroheme synthesis | | Descriptor: | PHOSPHATE ION, S-ADENOSYL-L-HOMOCYSTEINE, Siroheme synthase | | Authors: | Stroupe, M.E, Leech, H.K, Daniels, D.S, Warren, M.J, Getzoff, E.D. | | Deposit date: | 2003-06-03 | | Release date: | 2003-12-02 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | CysG structure reveals tetrapyrrole-binding features and novel regulation of siroheme biosynthesis.
Nat.Struct.Biol., 10, 2003
|
|
1OHL
 
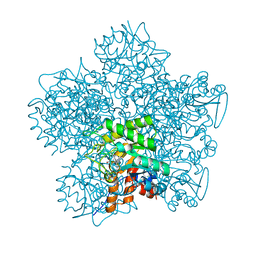 | | YEAST 5-AMINOLAEVULINIC ACID DEHYDRATASE PUTATIVE CYCLIC REACTION INTERMEDIATE COMPLEX | | Descriptor: | 3-[5-(AMINOMETHYL)-4-(CARBOXYMETHYL)-1H-PYRROL-3-YL]PROPANOIC ACID, BETA-MERCAPTOETHANOL, DELTA-AMINOLEVULINIC ACID DEHYDRATASE, ... | | Authors: | Erskine, P.T, Coates, L, Butler, D, Youell, J.H, Brindley, A.A, Wood, S.P, Warren, M.J, Shoolingin-Jordan, P.M, Cooper, J.B. | | Deposit date: | 2003-05-27 | | Release date: | 2003-06-02 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | X-Ray Structure of a Putative Reaction Intermediateof 5-Aminolaevulinic Acid Dehydratase
Biochem.J., 373, 2003
|
|
1QNV
 
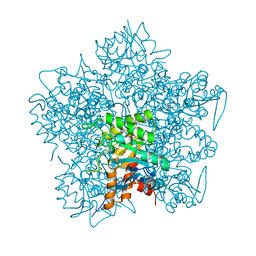 | | yeast 5-aminolaevulinic acid dehydratase Lead (Pb) complex | | Descriptor: | 5-AMINOLAEVULINIC ACID DEHYDRATASE, LEAD (II) ION | | Authors: | Erskine, P.T, Senior, N.M, Warren, M.J, Wood, S.P, Cooper, J.B. | | Deposit date: | 1999-10-21 | | Release date: | 2000-10-19 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | MAD Analyses of Yeast 5-Aminolaevulinic Acid Dehydratase. Their Use in Structure Determination and in Defining the Metal Binding Sites
Acta Crystallogr.,Sect.D, 56, 2000
|
|
1QML
 
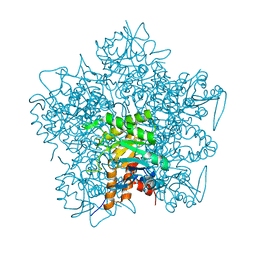 | | Hg complex of yeast 5-aminolaevulinic acid dehydratase | | Descriptor: | 5-AMINOLAEVULINIC ACID DEHYDRATASE, MERCURY (II) ION | | Authors: | Erskine, P.T, Senior, N, Warren, M.J, Wood, S.P, Cooper, J.B. | | Deposit date: | 1999-10-02 | | Release date: | 2000-10-06 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | MAD Analyses of Yeast 5-Aminolaevulinic Acid Dehydratase. Their Use in Structure Determination and in Defining the Metal Binding Sites
Acta Crystallogr.,Sect.D, 56, 2000
|
|
1EB3
 
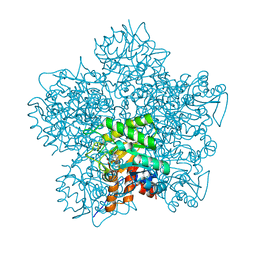 | | YEAST 5-AMINOLAEVULINIC ACID DEHYDRATASE 4,7-DIOXOSEBACIC ACID COMPLEX | | Descriptor: | 4,7-DIOXOSEBACIC ACID, 5-AMINOLAEVULINIC ACID DEHYDRATASE, ZINC ION | | Authors: | Erskine, P.T, Coates, L, Newbold, R, Brindley, A.A, Stauffer, F, Wood, S.P, Warren, M.J, Cooper, J.B, Shoolingin-Jordan, P.M, Neier, R. | | Deposit date: | 2001-07-18 | | Release date: | 2001-08-02 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | The X-Ray Structure of Yeast 5-Aminolaevulinic Acid Dehydratase Complexed with Two Diacid Inhibitors
FEBS Lett., 503, 2001
|
|
1S4D
 
 | | Crystal Structure Analysis of the S-adenosyl-L-methionine dependent uroporphyrinogen-III C-methyltransferase SUMT | | Descriptor: | GLYCEROL, S-ADENOSYL-L-HOMOCYSTEINE, Uroporphyrin-III C-methyltransferase | | Authors: | Vevodova, J, Graham, R.M, Raux, E, Schubert, H.L, Roper, D.I, Brindley, A.A, Scott, A.I, Roessner, C.A, Stamford, N.P.J, Stroupe, M.E, Getzoff, E.D, Warren, M.J, Wilson, K.S. | | Deposit date: | 2004-01-16 | | Release date: | 2004-11-30 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structure/Function Studies on a S-Adenosyl-l-methionine-dependent Uroporphyrinogen III C Methyltransferase (SUMT), a Key Regulatory Enzyme of Tetrapyrrole Biosynthesis
J.Mol.Biol., 344, 2004
|
|
3NJR
 
 | |
5C4R
 
 | | CobK precorrin-6A reductase | | Descriptor: | Precorrin-6A reductase | | Authors: | Gu, S, Pickersgill, R.W. | | Deposit date: | 2015-06-18 | | Release date: | 2016-11-16 | | Last modified: | 2017-08-30 | | Method: | X-RAY DIFFRACTION (3.17 Å) | | Cite: | Crystal structure of CobK reveals strand-swapping between Rossmann-fold domains and molecular basis of the reduced precorrin product trap.
Sci Rep, 5, 2015
|
|
3M4Z
 
 | | Crystal Structure of B. subtilis ferrochelatase with Cobalt bound at the active site | | Descriptor: | CHLORIDE ION, COBALT (II) ION, Ferrochelatase, ... | | Authors: | Soderberg, C.A.G, Hansson, M.D, Sreekanth, R, Al-Karadaghi, S, Hansson, M. | | Deposit date: | 2010-03-12 | | Release date: | 2010-11-10 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Bacterial ferrochelatase goes human: Tyr13 determines the apparent metal specificity of Bacillus subtilis ferrochelatase
To be Published
|
|
2XVY
 
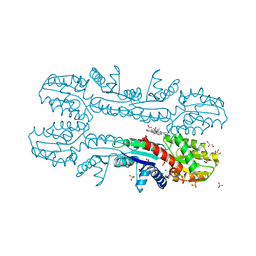 | | Cobalt chelatase CbiK (periplasmic) from Desulvobrio vulgaris Hildenborough (co-crystallised with cobalt and SHC) | | Descriptor: | CHELATASE, PUTATIVE, COBALT (II) ION, ... | | Authors: | Romao, C.V, Lobo, S.A.L, Carrondo, M.A, Saraiva, L.M, Matias, P.M. | | Deposit date: | 2010-10-28 | | Release date: | 2011-11-16 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Desulfovibrio vulgaris CbiK(P) cobaltochelatase: evolution of a haem binding protein orchestrated by the incorporation of two histidine residues.
Environ. Microbiol., 19, 2017
|
|
2XVZ
 
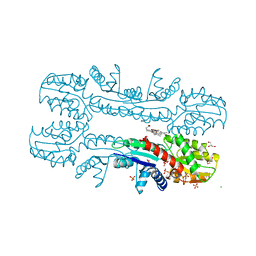 | | Cobalt chelatase CbiK (periplasmatic) from Desulvobrio vulgaris Hildenborough (co-crystallized with cobalt) | | Descriptor: | CHELATASE, PUTATIVE, CHLORIDE ION, ... | | Authors: | Romao, C.V, Lobo, S.A.L, Carrondo, M.A, Saraiva, L.M, Matias, P.M. | | Deposit date: | 2010-10-28 | | Release date: | 2010-12-22 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Evolution in a Family of Chelatases Facilitated by the Introduction of Active Site Asymmetry and Protein Oligomerization.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
2XVX
 
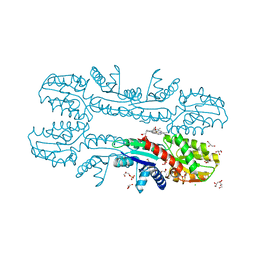 | | Cobalt chelatase CbiK (periplasmatic) from Desulvobrio vulgaris Hildenborough (Native) | | Descriptor: | CARBON DIOXIDE, CHELATASE, PUTATIVE, ... | | Authors: | Romao, C.V, Lobo, S.A.L, Carrondo, M.A, Saraiva, L.M, Matias, P.M. | | Deposit date: | 2010-10-28 | | Release date: | 2010-12-22 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Evolution in a Family of Chelatases Facilitated by the Introduction of Active Site Asymmetry and Protein Oligomerization.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
2DJ5
 
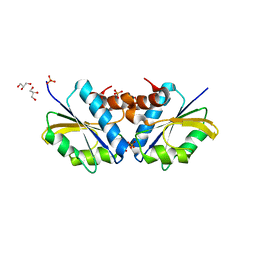 | | Crystal Structure of the vitamin B12 biosynthetic cobaltochelatase, CbiXS, from Archaeoglobus fulgidus | | Descriptor: | GLYCEROL, PHOSPHATE ION, Sirohydrochlorin cobaltochelatase | | Authors: | Yin, J, Cherney, M.M, James, M.N.G. | | Deposit date: | 2006-03-31 | | Release date: | 2006-09-12 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Crystal Structure of the Vitamin B(12) Biosynthetic Cobaltochelatase, CbiX (S), from Archaeoglobus Fulgidus
J.STRUCT.FUNCT.GENOM., 7, 2006
|
|
1AW5
 
 | |
