1OMY
 
 | | Crystal Structure of a Recombinant alpha-insect Toxin BmKaIT1 from the scorpion Buthus martensii Karsch | | Descriptor: | ACETIC ACID, Alpha-neurotoxin TX12, CHLORIDE ION | | Authors: | Huang, Y, Huang, Q, Chen, H, Tang, Y, Miyake, H, Kusunoki, M. | | Deposit date: | 2003-02-26 | | Release date: | 2003-09-09 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystallization and preliminary crystallographic study of rBmKalphaIT1, a recombinant alpha-insect toxin from the scorpion Buthus martensii Karsch.
Acta Crystallogr.,Sect.D, 59, 2003
|
|
3WJ1
 
 | | Crystal structure of SSHESTI | | Descriptor: | Carboxylesterase, octyl beta-D-glucopyranoside | | Authors: | Ohara, K, Unno, H, Oshima, Y, Furukawa, K, Fujino, N, Hirooka, K, Hemmi, H, Takahashi, S, Nishino, T, Kusunoki, M, Nakayama, T. | | Deposit date: | 2013-10-03 | | Release date: | 2014-07-30 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural insights into the low pH adaptation of a unique carboxylesterase from Ferroplasma: altering the pH optima of two carboxylesterases.
J.Biol.Chem., 289, 2014
|
|
1OX1
 
 | | crystal structure of the bovine trypsin complex with a synthetic 11 peptide inhibitor | | Descriptor: | 11-mer peptide, CALCIUM ION, Trypsinogen, ... | | Authors: | Wu, G, Huang, Y, Zhu, G, Huang, Q, Tang, Y, Miyake, H, Kusunoki, M. | | Deposit date: | 2003-03-31 | | Release date: | 2004-05-18 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | crystal structure of the bovine trypsin complex with a synthetic 11 peptide inhibitor
To be published
|
|
5GHL
 
 | | Crystal structure Analysis of the starch-binding domain of glucoamylase from Aspergillus niger | | Descriptor: | GLYCEROL, Glucoamylase, SULFATE ION | | Authors: | Miyake, H, Suyama, Y, Muraki, N, Kusunoki, M, Tanaka, A. | | Deposit date: | 2016-06-20 | | Release date: | 2017-10-18 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of the starch-binding domain of glucoamylase from Aspergillus niger.
Acta Crystallogr.,Sect.F, 73, 2017
|
|
1VCL
 
 | | Crystal Structure of Hemolytic Lectin CEL-III | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Uchida, T, Yamasaki, T, Eto, S, Sugawara, H, Kurisu, G, Nakagawa, A, Kusunoki, M, Hatakeyama, T. | | Deposit date: | 2004-03-09 | | Release date: | 2004-09-07 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal Structure of the Hemolytic Lectin CEL-III Isolated from the Marine Invertebrate Cucumaria echinata: IMPLICATIONS OF DOMAIN STRUCTURE FOR ITS MEMBRANE PORE-FORMATION MECHANISM
J.Biol.Chem., 279, 2004
|
|
1X1N
 
 | | Structure determination and refinement at 1.8 A resolution of Disproportionating Enzyme from Potato | | Descriptor: | 4-alpha-glucanotransferase, CALCIUM ION, GLYCEROL | | Authors: | Imamura, K, Matsuura, T, Takaha, T, Fujii, K, Nakagawa, A, Kusunoki, M, Nitta, Y. | | Deposit date: | 2005-04-08 | | Release date: | 2006-04-18 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure determination and refinement at 1.8 A resolution of Disproportionating Enzyme from Potato
to be published
|
|
3A28
 
 | | Crystal structure of L-2,3-butanediol dehydrogenase | | Descriptor: | BETA-MERCAPTOETHANOL, L-2.3-butanediol dehydrogenase, MAGNESIUM ION, ... | | Authors: | Otagiri, M, Kurisu, G, Ui, S, Kusunoki, M. | | Deposit date: | 2009-05-02 | | Release date: | 2009-12-15 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis for chiral substrate recognition by two 2,3-butanediol dehydrogenases
Febs Lett., 584, 2010
|
|
2E7P
 
 | | Crystal structure of the holo form of glutaredoxin C1 from populus tremula x tremuloides | | Descriptor: | FE2/S2 (INORGANIC) CLUSTER, GLUTATHIONE, Glutaredoxin | | Authors: | Unno, H, Takahashi, T, Kawakami, T, Aimoto, S, Hase, T, Kusunoki, M, Rouhier, N, Jacquot, J.P. | | Deposit date: | 2007-01-12 | | Release date: | 2007-09-25 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Functional, structural, and spectroscopic characterization of a glutathione-ligated [2Fe-2S] cluster in poplar glutaredoxin C1
Proc.Natl.Acad.Sci.Usa, 104, 2007
|
|
3WYE
 
 | | Crystal Structure of chimeric engineered (2S,3S)-butanediol dehydrogenase complexed with NAD+ | | Descriptor: | Diacetyl reductase [(S)-acetoin forming],L-2,3-butanediol dehydrogenase,Diacetyl reductase [(S)-acetoin forming],L-2,3-butanediol dehydrogenase,Diacetyl reductase [(S)-acetoin forming],L-2,3-butanediol dehydrogenase,Diacetyl reductase [(S)-acetoin forming], NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Shimegi, T, Oyama, T, Kusunoki, M, Ui, S. | | Deposit date: | 2014-08-26 | | Release date: | 2015-08-26 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Crystal Structure of chimeric engineered (2S,3S)-butanediol dehydrogenase complexed with NAD+
To be Published
|
|
1J18
 
 | | Crystal Structure of a Beta-Amylase from Bacillus cereus var. mycoides Cocrystallized with Maltose | | Descriptor: | ACETIC ACID, Beta-amylase, CALCIUM ION, ... | | Authors: | Miyake, H, Kurisu, G, Kusunoki, M, Nishimura, S, Kitamura, S, Nitta, Y. | | Deposit date: | 2002-12-02 | | Release date: | 2003-05-27 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of a Catalytic Site Mutant of beta-Amylase from Bacillus cereus var. mycoides Cocrystallized with Maltopentaose
BIOCHEMISTRY, 42, 2003
|
|
1ITC
 
 | | Beta-Amylase from Bacillus cereus var. mycoides Complexed with Maltopentaose | | Descriptor: | ACETIC ACID, Beta-Amylase, CALCIUM ION, ... | | Authors: | Miyake, H, Kurisu, G, Kusunoki, M, Nishimura, S, Kitamura, S, Nitta, Y. | | Deposit date: | 2002-01-17 | | Release date: | 2003-05-27 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structure of a Catalytic Site Mutant of beta-Amylase from Bacillus cereus var. mycoides Cocrystallized with Maltopentaose
BIOCHEMISTRY, 42, 2003
|
|
2D3C
 
 | | Crystal Structure of the Maize Glutamine Synthetase complexed with ADP and Phosphinothricin Phosphate | | Descriptor: | (2S)-2-AMINO-4-[METHYL(PHOSPHONOOXY)PHOSPHORYL]BUTANOIC ACID, ADENOSINE-5'-DIPHOSPHATE, MANGANESE (II) ION, ... | | Authors: | Unno, H, Uchida, T, Sugawara, H, Kurisu, G, Sugiyama, T, Yamaya, T, Sakakibara, H, Hase, T, Kusunoki, M. | | Deposit date: | 2005-09-26 | | Release date: | 2006-07-18 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (3.81 Å) | | Cite: | Atomic Structure of Plant Glutamine Synthetase: A KEY ENZYME FOR PLANT PRODUCTIVITY
J.Biol.Chem., 281, 2006
|
|
2E1U
 
 | |
2E1V
 
 | | Crystal structure of Dendranthema morifolium DmAT, seleno-methionine derivative | | Descriptor: | acyl transferase | | Authors: | Unno, H, Ichimaida, F, Kusunoki, M, Nakayama, T. | | Deposit date: | 2006-10-28 | | Release date: | 2007-04-10 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural and Mutational Studies of Anthocyanin Malonyltransferases Establish the Features of BAHD Enzyme Catalysis
J.Biol.Chem., 282, 2007
|
|
2E1T
 
 | | Crystal structure of Dendranthema morifolium DmAT complexed with malonyl-CoA | | Descriptor: | MALONYL-COENZYME A, acyl transferase | | Authors: | Unno, H, Ichimaida, F, Kusunoki, M, Nakayama, T. | | Deposit date: | 2006-10-28 | | Release date: | 2007-04-10 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural and Mutational Studies of Anthocyanin Malonyltransferases Establish the Features of BAHD Enzyme Catalysis
J.Biol.Chem., 282, 2007
|
|
2D3B
 
 | | Crystal Structure of the Maize Glutamine Synthetase complexed with AMPPNP and Methionine sulfoximine | | Descriptor: | (2S)-2-AMINO-4-(METHYLSULFONIMIDOYL)BUTANOIC ACID, MANGANESE (II) ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ... | | Authors: | Unno, H, Uchida, T, Sugawara, H, Kurisu, G, Sugiyama, T, Yamaya, T, Sakakibara, H, Hase, T, Kusunoki, M. | | Deposit date: | 2005-09-26 | | Release date: | 2006-07-18 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Atomic Structure of Plant Glutamine Synthetase: A KEY ENZYME FOR PLANT PRODUCTIVITY
J.Biol.Chem., 281, 2006
|
|
2D3A
 
 | | Crystal Structure of the Maize Glutamine Synthetase complexed with ADP and Methionine sulfoximine Phosphate | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, L-METHIONINE-S-SULFOXIMINE PHOSPHATE, MANGANESE (II) ION, ... | | Authors: | Unno, H, Uchida, T, Sugawara, H, Kurisu, G, Sugiyama, T, Yamaya, T, Sakakibara, H, Hase, T, Kusunoki, M. | | Deposit date: | 2005-09-26 | | Release date: | 2006-07-18 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.63 Å) | | Cite: | Atomic Structure of Plant Glutamine Synthetase: A KEY ENZYME FOR PLANT PRODUCTIVITY
J.Biol.Chem., 281, 2006
|
|
2ZOF
 
 | | Crystal structure of mouse carnosinase CN2 complexed with MN and bestatin | | Descriptor: | 2-(3-AMINO-2-HYDROXY-4-PHENYL-BUTYRYLAMINO)-4-METHYL-PENTANOIC ACID, Cytosolic non-specific dipeptidase, MANGANESE (II) ION | | Authors: | Unno, H, Yamashita, T, Okumura, N, Kusunoki, M. | | Deposit date: | 2008-05-14 | | Release date: | 2008-06-10 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural basis for substrate recognition and hydrolysis by mouse carnosinase CN2.
J.Biol.Chem., 283, 2008
|
|
2ZRV
 
 | | Crystal structure of Sulfolobus shibatae isopentenyl diphosphate isomerase in complex with reduced FMN. | | Descriptor: | 1-DEOXY-1-(7,8-DIMETHYL-2,4-DIOXO-3,4-DIHYDRO-2H-BENZO[G]PTERIDIN-1-ID-10(5H)-YL)-5-O-PHOSPHONATO-D-RIBITOL, Isopentenyl-diphosphate delta-isomerase | | Authors: | Unno, H, Yamashita, S, Ikeda, Y, Sekiguchi, S, Yoshida, N, Yoshimura, T, Kusunoki, M, Nakayama, T, Nishino, T, Hemmi, H. | | Deposit date: | 2008-09-01 | | Release date: | 2009-01-20 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | New role of flavin as a general acid-base catalyst with no redox function in type 2 isopentenyl-diphosphate isomerase.
J.Biol.Chem., 284, 2009
|
|
2ZRW
 
 | | Crystal structure of Sulfolobus shibatae isopentenyl diphosphate isomerase in complex with FMN and IPP. | | Descriptor: | FLAVIN MONONUCLEOTIDE, ISOPENTYL PYROPHOSPHATE, Isopentenyl-diphosphate delta-isomerase, ... | | Authors: | Unno, H, Yamashita, S, Ikeda, Y, Sekiguchi, S, Yoshida, N, Yoshimura, T, Kusunoki, M, Nakayama, T, Nishino, T, Hemmi, H. | | Deposit date: | 2008-09-01 | | Release date: | 2009-01-20 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | New role of flavin as a general acid-base catalyst with no redox function in type 2 isopentenyl-diphosphate isomerase.
J.Biol.Chem., 284, 2009
|
|
3A4A
 
 | | Crystal structure of isomaltase from Saccharomyces cerevisiae | | Descriptor: | CALCIUM ION, Oligo-1,6-glucosidase, alpha-D-glucopyranose | | Authors: | Yamamoto, K, Miyake, H, Kusunoki, M, Osaki, S. | | Deposit date: | 2009-07-01 | | Release date: | 2010-07-14 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structures of isomaltase from Saccharomyces cerevisiae and in complex with its competitive inhibitor maltose
Febs J., 277, 2010
|
|
3A47
 
 | |
3ATW
 
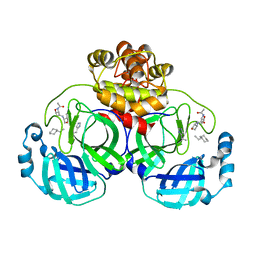 | | Structure-Based Design, Synthesis, Evaluation of Peptide-mimetic SARS 3CL Protease Inhibitors | | Descriptor: | 3C-Like Proteinase, peptide ACE-THR-VAL-ALC-HIS-H | | Authors: | Akaji, K, Konno, H, Mitsui, H, Teruya, K, Hattori, Y, Ozaki, T, Kusunoki, M, Sanjho, A. | | Deposit date: | 2011-01-20 | | Release date: | 2011-12-14 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.36 Å) | | Cite: | Structure-Based Design, Synthesis, and Evaluation of Peptide-Mimetic SARS 3CL Protease Inhibitors.
J.Med.Chem., 54, 2011
|
|
3AVZ
 
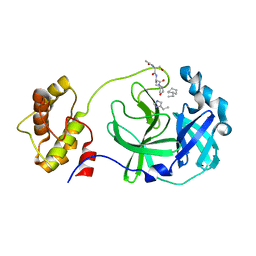 | | Structure of SARS 3CL protease with peptidic aldehyde inhibitor containing cyclohexyl side chain | | Descriptor: | 3C-Like Proteinase, peptide ACE-SER-ALA-VAL-ALC-HIS-H | | Authors: | Akaji, K, Konno, H, Mitsui, H, Teruya, K, Hattori, Y, Ozaki, T, Kusunoki, M, Sanjho, A. | | Deposit date: | 2011-03-09 | | Release date: | 2011-12-14 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.46 Å) | | Cite: | Structure-Based Design, Synthesis, and Evaluation of Peptide-Mimetic SARS 3CL Protease Inhibitors.
J.Med.Chem., 54, 2011
|
|
3AXH
 
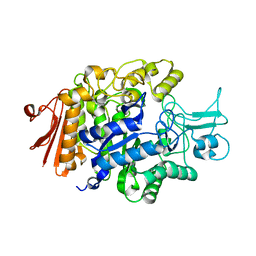 | | Crystal structure of isomaltase in complex with isomaltose | | Descriptor: | CALCIUM ION, Oligo-1,6-glucosidase IMA1, alpha-D-glucopyranose-(1-6)-alpha-D-glucopyranose | | Authors: | Yamamoto, K, Miyake, H, Kusunoki, M, Osaki, S. | | Deposit date: | 2011-04-06 | | Release date: | 2011-10-05 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Steric hindrance by 2 amino acid residues determines the substrate specificity of isomaltase from Saccharomyces cerevisiae
J.Biosci.Bioeng., 112, 2011
|
|
