7M7B
 
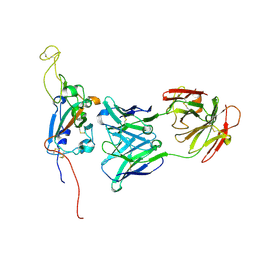 | | SARS-CoV-2 Spike:Fab 3D11 complex focused refinement | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Antibody Fab 3D11 heavy chain, Antibody Fab 3D11 light chain, ... | | Authors: | Asarnow, D, Cheng, Y. | | Deposit date: | 2021-03-27 | | Release date: | 2021-05-26 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (2.95 Å) | | Cite: | Structural insight into SARS-CoV-2 neutralizing antibodies and modulation of syncytia.
Cell, 184, 2021
|
|
4UG1
 
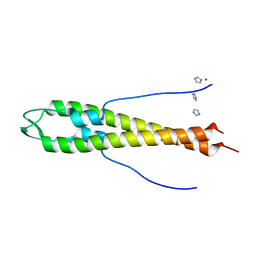 | | GpsB N-terminal domain | | Descriptor: | CELL CYCLE PROTEIN GPSB, IMIDAZOLE, NICKEL (II) ION | | Authors: | Rismondo, J, Cleverley, R.M, Lane, H.V, Grohennig, S, Steglich, A, Muller, L, Krishna Mannala, G, Hain, T, Lewis, R.J, Halbedel, S. | | Deposit date: | 2015-03-20 | | Release date: | 2015-11-25 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure of the Bacterial Cell Division Determinant Gpsb and its Interaction with Penicillin Binding Proteins.
Mol.Microbiol., 99, 2016
|
|
4UG3
 
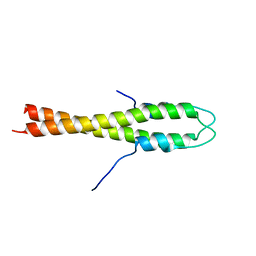 | | B. subtilis GpsB N-terminal Domain | | Descriptor: | CELL CYCLE PROTEIN GPSB | | Authors: | Rismondo, J, Cleverley, R.M, Lane, H.V, Grohennig, S, Steglich, A, Moller, L, Krishna Mannala, G, Hain, T, Lewis, R.J, Halbedel, S. | | Deposit date: | 2015-03-21 | | Release date: | 2015-11-25 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of the Bacterial Cell Division Determinant Gpsb and its Interaction with Penicillin Binding Proteins.
Mol.Microbiol., 99, 2016
|
|
1R8Y
 
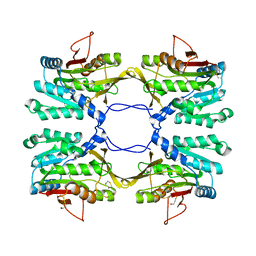 | | Crystal Structure of Mouse Glycine N-Methyltransferase (Monoclinic Form) | | Descriptor: | BETA-MERCAPTOETHANOL, glycine N-methyltransferase | | Authors: | Pakhomova, S, Luka, Z, Wagner, C, Newcomer, M.E. | | Deposit date: | 2003-10-28 | | Release date: | 2004-09-21 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Glycine N-methyltransferases: a comparison of the crystal structures and kinetic properties of recombinant human, mouse and rat enzymes.
Proteins, 57, 2004
|
|
6N6B
 
 | |
1R74
 
 | | Crystal Structure of Human Glycine N-Methyltransferase | | Descriptor: | BETA-MERCAPTOETHANOL, CITRIC ACID, Glycine N-methyltransferase | | Authors: | Pakhomova, S, Luka, Z, Wagner, C, Newcomer, M.E. | | Deposit date: | 2003-10-17 | | Release date: | 2004-09-21 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Glycine N-methyltransferases: a comparison of the crystal structures and kinetic properties of recombinant human, mouse and rat enzymes.
Proteins, 57, 2004
|
|
1R8X
 
 | | Crystal Structure of Mouse Glycine N-Methyltransferase (Tetragonal Form) | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, BETA-MERCAPTOETHANOL, glycine N-methyltransferase | | Authors: | Pakhomova, S, Luka, Z, Wagner, C, Newcomer, M.E. | | Deposit date: | 2003-10-28 | | Release date: | 2004-09-21 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Glycine N-methyltransferases: a comparison of the crystal structures and kinetic properties of recombinant human, mouse and rat enzymes.
Proteins, 57, 2004
|
|
5LAR
 
 | | Crystal structure of p38 alpha MAPK14 in complex with VPC00628 | | Descriptor: | 5-azanyl-~{N}-[[4-[[(2~{S})-1-azanyl-4-cyclohexyl-1-oxidanylidene-butan-2-yl]carbamoyl]phenyl]methyl]-1-phenyl-pyrazole-4-carboxamide, Mitogen-activated protein kinase 14 | | Authors: | Chaikuad, A, Petersen, L.K, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2016-06-14 | | Release date: | 2016-07-06 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Novel p38alpha MAP kinase inhibitors identified from yoctoReactor DNA-encoded small molecule library
Medchemcomm, 7, 2016
|
|
5AN5
 
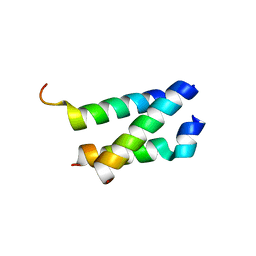 | | B. subtilis GpsB C-terminal Domain | | Descriptor: | CELL CYCLE PROTEIN GPSB, GLYCEROL | | Authors: | Rismondo, J, Cleverley, R.M, Lane, H.V, Grohennig, S, Steglich, A, Moller, L, Krishna Mannala, G, Hain, T, Lewis, R.J, Halbedel, S. | | Deposit date: | 2015-09-04 | | Release date: | 2015-11-25 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Structure of the Bacterial Cell Division Determinant Gpsb and its Interaction with Penicillin Binding Proteins.
Mol.Microbiol., 99, 2016
|
|
9FJD
 
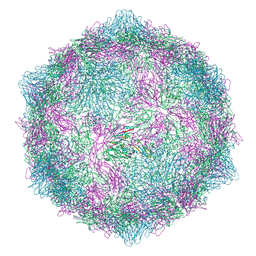 | | Expanded CVB1-VLP (Tween80) | | Descriptor: | Capsid protein VP1, Capsid protein VP2, Capsid protein VP3 | | Authors: | Plavec, Z, Butcher, S.J. | | Deposit date: | 2024-05-31 | | Release date: | 2024-07-31 | | Method: | ELECTRON MICROSCOPY (2.15 Å) | | Cite: | Comparison of structure and immunogenicity of CVB1-VLP and inactivated CVB1 vaccine candidates.
Res Sq, 2024
|
|
9FJE
 
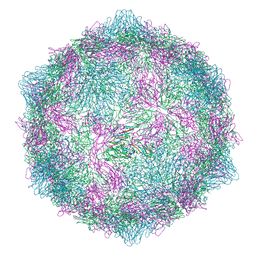 | | Expanded formalin inactivated CVB1 | | Descriptor: | Capsid protein VP1, Capsid protein VP2, Capsid protein VP3 | | Authors: | Plavec, Z, Butcher, S.J. | | Deposit date: | 2024-05-31 | | Release date: | 2024-07-31 | | Method: | ELECTRON MICROSCOPY (3.01 Å) | | Cite: | Comparison of structure and immunogenicity of CVB1-VLP and inactivated CVB1 vaccine candidates.
Res Sq, 2024
|
|
9FJC
 
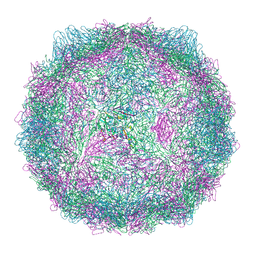 | | Compact CVB1-VLP (Tween80) | | Descriptor: | Capsid protein VP1, Capsid protein VP2, Capsid protein VP3, ... | | Authors: | Plavec, Z, Butcher, S.J. | | Deposit date: | 2024-05-30 | | Release date: | 2024-07-31 | | Method: | ELECTRON MICROSCOPY (2.15 Å) | | Cite: | Comparison of structure and immunogenicity of CVB1-VLP and inactivated CVB1 vaccine candidates.
Res Sq, 2024
|
|
9FZA
 
 | |
6ZVQ
 
 | | Complex between SMAD2 MH2 domain and peptide from Ski corepressor | | Descriptor: | D(-)-TARTARIC ACID, GLYCEROL, Mothers against decapentaplegic homolog 2, ... | | Authors: | Purkiss, A.G, Kjaer, S, George, R, Hill, C.S. | | Deposit date: | 2020-07-27 | | Release date: | 2021-06-09 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Mutations in SKI in Shprintzen-Goldberg syndrome lead to attenuated TGF-beta responses through SKI stabilization.
Elife, 10, 2021
|
|
5NZU
 
 | | The structure of the COPI coat linkage II | | Descriptor: | ADP-ribosylation factor 1, Coatomer subunit alpha, Coatomer subunit beta, ... | | Authors: | Dodonova, S.O, Aderhold, P, Kopp, J, Ganeva, I, Roehling, S, Hagen, W.J.H, Sinning, I, Wieland, F, Briggs, J.A.G. | | Deposit date: | 2017-05-15 | | Release date: | 2017-06-28 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (15 Å) | | Cite: | 9 angstrom structure of the COPI coat reveals that the Arf1 GTPase occupies two contrasting molecular environments.
Elife, 6, 2017
|
|
5NZS
 
 | | The structure of the COPI coat leaf in complex with the ArfGAP2 uncoating factor | | Descriptor: | ADP-ribosylation factor 1, ADP-ribosylation factor GTPase-activating protein 2, Coatomer subunit alpha, ... | | Authors: | Dodonova, S.O, Aderhold, P, Kopp, J, Ganeva, I, Roehling, S, Hagen, W.J.H, Sinning, I, Wieland, F, Briggs, J.A.G. | | Deposit date: | 2017-05-15 | | Release date: | 2017-06-28 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (10.1 Å) | | Cite: | 9 angstrom structure of the COPI coat reveals that the Arf1 GTPase occupies two contrasting molecular environments.
Elife, 6, 2017
|
|
5BJT
 
 | |
5NZR
 
 | | The structure of the COPI coat leaf | | Descriptor: | ADP-ribosylation factor 1, Coatomer subunit alpha, Coatomer subunit beta, ... | | Authors: | Dodonova, S.O, Aderhold, P, Kopp, J, Ganeva, I, Roehling, S, Hagen, W.J.H, Sinning, I, Wieland, F, Briggs, J.A.G. | | Deposit date: | 2017-05-15 | | Release date: | 2017-06-28 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (9.2 Å) | | Cite: | 9 angstrom structure of the COPI coat reveals that the Arf1 GTPase occupies two contrasting molecular environments.
Elife, 6, 2017
|
|
5NZV
 
 | | The structure of the COPI coat linkage IV | | Descriptor: | ADP-ribosylation factor 1, Coatomer subunit alpha, Coatomer subunit beta, ... | | Authors: | Dodonova, S.O, Aderhold, P, Kopp, J, Ganeva, I, Roehling, S, Hagen, W.J.H, Sinning, I, Wieland, F, Briggs, J.A.G. | | Deposit date: | 2017-05-15 | | Release date: | 2017-06-28 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (17.299999 Å) | | Cite: | 9 angstrom structure of the COPI coat reveals that the Arf1 GTPase occupies two contrasting molecular environments.
Elife, 6, 2017
|
|
5NZT
 
 | | The structure of the COPI coat linkage I | | Descriptor: | ADP-ribosylation factor 1, Coatomer subunit alpha, Coatomer subunit beta, ... | | Authors: | Dodonova, S.O, Aderhold, P, Kopp, J, Ganeva, I, Roehling, S, Hagen, W.J.H, Sinning, I, Wieland, F, Briggs, J.A.G. | | Deposit date: | 2017-05-15 | | Release date: | 2017-06-28 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (17 Å) | | Cite: | 9 angstrom structure of the COPI coat reveals that the Arf1 GTPase occupies two contrasting molecular environments.
Elife, 6, 2017
|
|
7SFJ
 
 | | ChRmine in MSP1E3D1 lipid nanodisc | | Descriptor: | 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine, ChRmine, RETINAL | | Authors: | Tucker, K, Brohawn, S. | | Deposit date: | 2021-10-04 | | Release date: | 2021-12-01 | | Last modified: | 2022-08-31 | | Method: | ELECTRON MICROSCOPY (2.74 Å) | | Cite: | Cryo-EM structures of the channelrhodopsin ChRmine in lipid nanodiscs.
Nat Commun, 13, 2022
|
|
7SHS
 
 | |
7SFK
 
 | | ChRmine in MSP1E3D1 lipid nanodisc | | Descriptor: | 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine, ChRmine, RETINAL | | Authors: | Tucker, K, Brohawn, S. | | Deposit date: | 2021-10-04 | | Release date: | 2021-12-01 | | Last modified: | 2022-08-31 | | Method: | ELECTRON MICROSCOPY (2.74 Å) | | Cite: | Cryo-EM structures of the channelrhodopsin ChRmine in lipid nanodiscs.
Nat Commun, 13, 2022
|
|
6Q1Z
 
 | |
1NMK
 
 | | The Sanglifehrin-Cyclophilin Interaction: Degradation Work, Synthetic Macrocyclic Analogues, X-ray Crystal Structure and Binding Data | | Descriptor: | (13E,15E)-(3S,6S,9R,10R,11S,12S,18S,21S)-10,12-DIHYDROXY-3-(3-HYDROXYBEN-ZYL)-18-((E)-3-HYDROXY-1-METHYLPROPENYL)-6-ISOPROPYL-11-METHYL-9-(3-OXO-BUTYL)-19-OXA-1,4,7,25-TETRAAZA-BICYCLO[19.3.1]PENTACOSA-13,15-DIENE-2,5,8,20-TETRAONE, Peptidyl-prolyl cis-trans isomerase A | | Authors: | Kallen, J, Sedrani, R, Wagner, J. | | Deposit date: | 2003-01-10 | | Release date: | 2003-04-01 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Sanglifehrin-Cyclophilin Interaction: Degradation Work, Synthetic Macrocyclic Analogues, X-ray Crystal Structure and Binding Data
J.Am.Chem.Soc., 125, 2003
|
|
