6UBO
 
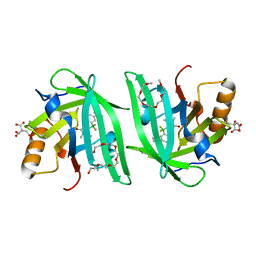 | | Fluorogen Activating Protein Dib1 | | Descriptor: | 12-(diethylamino)-2,2-bis(fluoranyl)-4,5-dimethyl-5-aza-3-azonia-2-boranuidatricyclo[7.4.0.0^{3,7}]trideca-1(13),3,7,9,11-pentaen-6-one, CITRIC ACID, Outer membrane lipoprotein Blc, ... | | Authors: | Muslinkina, L, Pletneva, N, Pletnev, V.Z, Pletnev, S. | | Deposit date: | 2019-09-12 | | Release date: | 2020-09-16 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Structure-Based Rational Design of Two Enhanced Bacterial Lipocalin Blc Tags for Protein-PAINT Super-resolution Microscopy.
Acs Chem.Biol., 15, 2020
|
|
399D
 
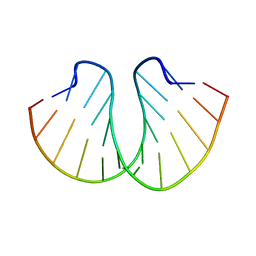 | | STRUCTURE OF D(CGCCCGCGGGCG) | | Descriptor: | DNA (5'-D(*CP*GP*CP*CP*CP*GP*CP*GP*GP*GP*CP*G)-3') | | Authors: | Malinina, L, Fernandez, L.G, Subirana, J.A. | | Deposit date: | 1998-05-11 | | Release date: | 1999-01-18 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of the d(CGCCCGCGGGCG) dodecamer: a kinked A-DNA molecule showing some B-DNA features.
J.Mol.Biol., 285, 1999
|
|
1YOE
 
 | |
1G3X
 
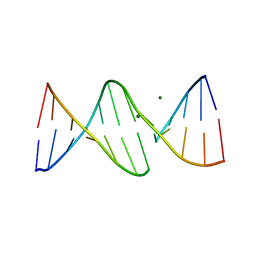 | | INTERCALATION OF AN 9ACRIDINE-PEPTIDE DRUG IN A DNA DODECAMER | | Descriptor: | 5'-D(*CP*GP*CP*GP*AP*AP*TP*TP*CP*GP*CP*G)-3', MAGNESIUM ION, N(ALPHA)-(9-ACRIDINOYL)-TETRAARGININE-AMIDE | | Authors: | Malinina, L, Soler-Lopez, M, Aymami, J, Subirana, J.A. | | Deposit date: | 2000-10-25 | | Release date: | 2002-08-16 | | Last modified: | 2024-07-10 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Intercalation of an acridine-peptide drug in an AA/TT base step in the crystal structure of [d(CGCGAATTCGCG)](2) with six duplexes and seven Mg(2+) ions in the asymmetric unit.
Biochemistry, 41, 2002
|
|
2EVD
 
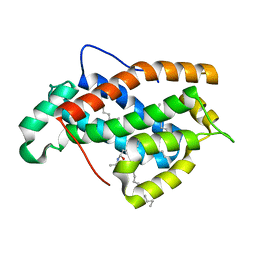 | | Crystal structure of human Glycolipid Transfer Protein complexed with 12:0 Lactosylceramide | | Descriptor: | DECANE, Glycolipid transfer protein, LAURIC ACID, ... | | Authors: | Malinina, L, Malakhova, M.L, Kanack, A.T, Abagyan, R, Brown, R.E, Patel, D.J. | | Deposit date: | 2005-10-31 | | Release date: | 2006-11-14 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The liganding of glycolipid transfer protein is controlled by glycolipid acyl structure.
Plos Biol., 4, 2006
|
|
2EVL
 
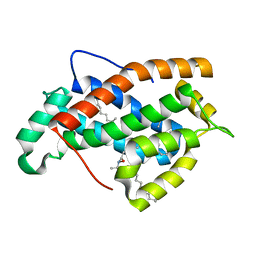 | | Crystal structure of human Glycolipid Transfer Protein complexed with 18:2 Galactosylceramide | | Descriptor: | Glycolipid transfer protein, LINOLEIC ACID, N-OCTANE, ... | | Authors: | Malinina, L, Malakhova, M.L, Kanack, A.T, Abagyan, R, Brown, R.E, Patel, D.J. | | Deposit date: | 2005-10-31 | | Release date: | 2006-11-14 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The liganding of glycolipid transfer protein is controlled by glycolipid acyl structure.
Plos Biol., 4, 2006
|
|
2EUM
 
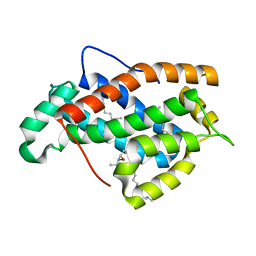 | | Crystal structure of human Glycolipid Transfer Protein complexed with 8:0 Lactosylceramide | | Descriptor: | DECANE, Glycolipid transfer protein, N-OCTANE, ... | | Authors: | Malinina, L, Malakhova, M.L, Kanack, A.T, Abagyan, R, Brown, R.E, Patel, D.J. | | Deposit date: | 2005-10-28 | | Release date: | 2006-11-14 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The liganding of glycolipid transfer protein is controlled by glycolipid acyl structure.
Plos Biol., 4, 2006
|
|
2EUK
 
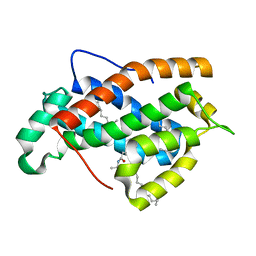 | | Crystal Structure of Human Glycolipid Transfer Protein complexed with 24:1 Galactosylceramide | | Descriptor: | (15E)-TETRACOS-15-ENOIC ACID, Glycolipid transfer protein, N-OCTANE, ... | | Authors: | Malinina, L, Malakhova, M.L, Kanack, A.T, Abagyan, R, Brown, R.E, Patel, D.J. | | Deposit date: | 2005-10-28 | | Release date: | 2006-11-14 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | The liganding of glycolipid transfer protein is controlled by glycolipid acyl structure.
Plos Biol., 4, 2006
|
|
2EVT
 
 | | Crystal structure of D48V mutant of human Glycolipid Transfer Protein | | Descriptor: | Glycolipid transfer protein, HEXANE | | Authors: | Malinina, L, Malakhova, M.L, Teplov, A, Brown, R.E, Patel, D.J. | | Deposit date: | 2005-10-31 | | Release date: | 2005-11-15 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | The liganding of glycolipid transfer protein is controlled by glycolipid acyl structure.
Plos Biol., 4, 2006
|
|
2EVS
 
 | | Crystal structure of human Glycolipid Transfer Protein complexed with n-hexyl-beta-D-glucoside | | Descriptor: | DECANE, Glycolipid transfer protein, HEXANE, ... | | Authors: | Malinina, L, Malakhova, M.L, Kanack, A.T, Abagyan, R, Brown, R.E, Patel, D.J. | | Deposit date: | 2005-10-31 | | Release date: | 2006-11-14 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The liganding of glycolipid transfer protein is controlled by glycolipid acyl structure.
Plos Biol., 4, 2006
|
|
1IL2
 
 | | Crystal Structure of the E. coli Aspartyl-tRNA Synthetase:Yeast tRNAasp:aspartyl-Adenylate Complex | | Descriptor: | ASPARTYL TRANSFER RNA, ASPARTYL-ADENOSINE-5'-MONOPHOSPHATE, ASPARTYL-TRNA SYNTHETASE, ... | | Authors: | Moulinier, L, Eiler, S, Eriani, G, Gangloff, J, Thierry, J.C, Gabriel, K, McClain, W.H, Moras, D. | | Deposit date: | 2001-05-07 | | Release date: | 2001-09-28 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The structure of an AspRS-tRNA(Asp) complex reveals a tRNA-dependent control mechanism.
EMBO J., 20, 2001
|
|
1AZR
 
 | | CRYSTAL STRUCTURE OF PSEUDOMONAS AERUGINOSA ZINC AZURIN MUTANT ASP47ASP AT 2.4 ANGSTROMS RESOLUTION | | Descriptor: | AZURIN, COPPER (II) ION, NITRATE ION | | Authors: | Sjolin, L, Tsai, Lc, Langer, V, Pascher, T, Karlsson, G, Nordling, M, Nar, H. | | Deposit date: | 1993-03-04 | | Release date: | 1993-07-15 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure of Pseudomonas aeruginosai zinc azurin mutant Asn47Asp at 2.4 A resolution.
Acta Crystallogr.,Sect.D, 49, 1993
|
|
192D
 
 | | RECOMBINATION-LIKE STRUCTURE OF D(CCGCGG) | | Descriptor: | DNA (5'-D(*CP*CP*GP*CP*GP*G)-3'), SODIUM ION | | Authors: | Malinina, L, Urpi, L, Salas, X, Huynh-Dinh, T, Subirana, J.A. | | Deposit date: | 1994-09-22 | | Release date: | 1995-02-07 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Recombination-like structure of d(CCGCGG).
J.Mol.Biol., 243, 1994
|
|
7BYJ
 
 | | Crystal structure of the FERM domain of FRMPD4 | | Descriptor: | FERM and PDZ domain-containing protein 4 | | Authors: | Lin, L, Wang, M, Wang, C, Zhu, J. | | Deposit date: | 2020-04-23 | | Release date: | 2020-12-16 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | Structure of the FERM domain of a neural scaffold protein FRMPD4 implicated in X-linked intellectual disability.
Biochem.J., 477, 2020
|
|
2ANN
 
 | | Crystal structure (I) of Nova-1 KH1/KH2 domain tandem with 25 nt RNA hairpin | | Descriptor: | 5'-R(*CP*GP*CP*GP*CP*GP*GP*AP*UP*CP*AP*GP*UP*CP*AP*CP*CP*CP*AP*AP*GP*CP*GP*CP*G)-3', MAGNESIUM ION, POTASSIUM ION, ... | | Authors: | Malinina, L, Teplova, M, Musunuru, K, Teplov, A, Darnell, J.C, Burley, S.K, Darnell, R.B, Patel, D.J. | | Deposit date: | 2005-08-11 | | Release date: | 2006-10-24 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Protein-RNA and protein-protein recognition by dual KH1/2 domains of the neuronal splicing factor Nova-1.
Structure, 19, 2011
|
|
5YMR
 
 | | The Crystal Structure of IseG | | Descriptor: | 2-hydroxyethylsulfonic acid, Formate acetyltransferase, GLYCEROL | | Authors: | Lin, L, Zhang, J, Xing, M, Hua, G, Guo, C, Hu, Y, Wei, Y, Ang, E, Zhao, H, Zhang, Y, Yuchi, Z. | | Deposit date: | 2017-10-22 | | Release date: | 2019-03-20 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Radical-mediated C-S bond cleavage in C2 sulfonate degradation by anaerobic bacteria.
Nat Commun, 10, 2019
|
|
6JKM
 
 | | Crystal structure of BubR1 kinase domain | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Lin, L, Ye, S, Huang, Y, Liu, X, Zhang, R, Yao, X. | | Deposit date: | 2019-03-01 | | Release date: | 2019-06-26 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | BubR1 phosphorylates CENP-E as a switch enabling the transition from lateral association to end-on capture of spindle microtubules.
Cell Res., 29, 2019
|
|
6JKK
 
 | | Crystal structure of BubR1 kinase domain | | Descriptor: | DI(HYDROXYETHYL)ETHER, GLYCEROL, Mitotic checkpoint control protein kinase BUB1 | | Authors: | Lin, L, Ye, S, Huang, Y, Liu, X, Zhang, R, Yao, X. | | Deposit date: | 2019-03-01 | | Release date: | 2019-06-26 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | BubR1 phosphorylates CENP-E as a switch enabling the transition from lateral association to end-on capture of spindle microtubules.
Cell Res., 29, 2019
|
|
6CW0
 
 | | Crystal structure of Cryptosporidium parvum bromodomain cgd2_2690 | | Descriptor: | 4-{[(7R)-8-cyclopentyl-7-ethyl-5-methyl-6-oxo-5,6,7,8-tetrahydropteridin-2-yl]amino}-3-methoxy-N-(1-methylpiperidin-4-yl)benzamide, Cgd2_2690 protein, GLYCEROL, ... | | Authors: | Dong, A, Lin, L, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Hui, R, Structural Genomics Consortium (SGC) | | Deposit date: | 2018-03-29 | | Release date: | 2018-04-18 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.38 Å) | | Cite: | Crystal structure of Cryptosporidium parvum bromodomain cgd2_2690
to be published
|
|
7DE7
 
 | | Crystal structure of PDZD7 HHD domain | | Descriptor: | (2R)-2-{[(2R)-2-{[(2S)-2-{[(2R)-2-hydroxypropyl]oxy}propyl]oxy}propyl]oxy}propan-1-ol, PDZ domain-containing protein 7 | | Authors: | Wang, H, Lin, L, Lu, Q. | | Deposit date: | 2020-11-02 | | Release date: | 2021-08-25 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Structure and Membrane Targeting of the PDZD7 Harmonin Homology Domain (HHD) Associated With Hearing Loss.
Front Cell Dev Biol, 9, 2021
|
|
7EP7
 
 | | The complex structure of Gpsm2 and Whirlin | | Descriptor: | G-protein-signaling modulator 2, Whirlin | | Authors: | Lin, L, Shi, Y, Wang, C, Zhu, J. | | Deposit date: | 2021-04-26 | | Release date: | 2022-05-04 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Promotion of row 1-specific tip complex condensates by Gpsm2-G alpha i provides insights into row identity of the tallest stereocilia.
Sci Adv, 8, 2022
|
|
6YPO
 
 | | Arabidopsis aspartate transcarbamoylase bound to UMP | | Descriptor: | GLYCEROL, PYRB, URIDINE-5'-MONOPHOSPHATE | | Authors: | Ramon Maiques, S, Del Cano Ochoa, F, Bellin, L, Mohlmann, T. | | Deposit date: | 2020-04-16 | | Release date: | 2021-03-03 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Mechanisms of feedback inhibition and sequential firing of active sites in plant aspartate transcarbamoylase.
Nat Commun, 12, 2021
|
|
6YS6
 
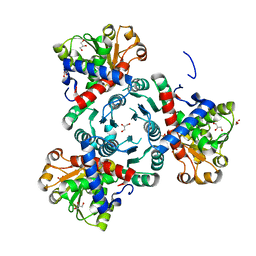 | | Arabidopsis aspartate transcarbamoylase complex with PALA | | Descriptor: | GLYCEROL, N-(PHOSPHONACETYL)-L-ASPARTIC ACID, PYRB, ... | | Authors: | Ramon Maiques, S, Del Cano Ochoa, F, Bellin, L, Mohlmann, T. | | Deposit date: | 2020-04-21 | | Release date: | 2021-03-03 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Mechanisms of feedback inhibition and sequential firing of active sites in plant aspartate transcarbamoylase.
Nat Commun, 12, 2021
|
|
6YSP
 
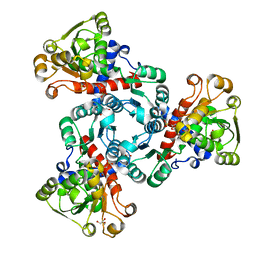 | | Arabidopsis aspartate transcarbamoylase complex with PALA and carbamoyl phosphate | | Descriptor: | GLYCEROL, N-(PHOSPHONACETYL)-L-ASPARTIC ACID, PHOSPHORIC ACID MONO(FORMAMIDE)ESTER, ... | | Authors: | Ramon Maiques, S, Del Cano Ochoa, F, Bellin, L, Mohlmann, T. | | Deposit date: | 2020-04-23 | | Release date: | 2021-03-03 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.38 Å) | | Cite: | Mechanisms of feedback inhibition and sequential firing of active sites in plant aspartate transcarbamoylase.
Nat Commun, 12, 2021
|
|
6YW9
 
 | | Arabidopsis aspartate transcarbamoylase mutant F161A complex with PALA | | Descriptor: | GLYCEROL, N-(PHOSPHONACETYL)-L-ASPARTIC ACID, PYRB | | Authors: | Ramon Maiques, S, Del Cano Ochoa, F, Bellin, L, Mohlmann, T. | | Deposit date: | 2020-04-29 | | Release date: | 2021-03-03 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Mechanisms of feedback inhibition and sequential firing of active sites in plant aspartate transcarbamoylase.
Nat Commun, 12, 2021
|
|
