6XA9
 
 | | SARS CoV-2 PLpro in complex with ISG15 C-terminal domain propargylamide | | Descriptor: | GLYCEROL, ISG15 CTD-propargylamide, Non-structural protein 3, ... | | Authors: | Klemm, T, Calleja, D.J, Richardson, L.W, Lechtenberg, B.C, Komander, D. | | Deposit date: | 2020-06-04 | | Release date: | 2020-06-17 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Mechanism and inhibition of the papain-like protease, PLpro, of SARS-CoV-2.
Embo J., 39, 2020
|
|
6XAA
 
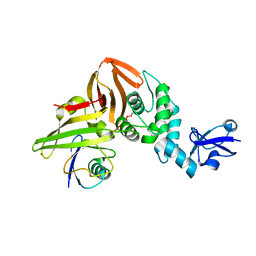 | | SARS CoV-2 PLpro in complex with ubiquitin propargylamide | | Descriptor: | Non-structural protein 3, Ubiquitin-propargylamide, ZINC ION | | Authors: | Klemm, T, Calleja, D.J, Richardson, L.W, Lechtenberg, B.C, Komander, D. | | Deposit date: | 2020-06-04 | | Release date: | 2020-06-17 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Mechanism and inhibition of the papain-like protease, PLpro, of SARS-CoV-2.
Embo J., 39, 2020
|
|
2VYT
 
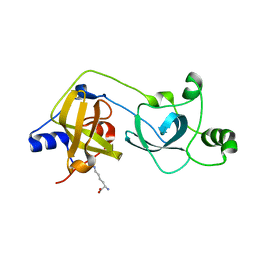 | | The MBT repeats of human SCML2 bind to peptides containing mono methylated lysine. | | Descriptor: | N-METHYL-LYSINE, SEX COMB ON MIDLEG-LIKE PROTEIN 2, TRIETHYLENE GLYCOL | | Authors: | Santiveri, C.M, Lechtenberg, B.C, Allen, M.D, Sathyamurthy, A, Jaulent, A.M, Freund, S.M.V, Bycroft, M. | | Deposit date: | 2008-07-28 | | Release date: | 2008-08-05 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The Malignant Brain Tumor Repeats of Human Scml2 Bind to Peptides Containing Monomethylated Lysine.
J.Mol.Biol., 382, 2008
|
|
7M4O
 
 | |
7M4M
 
 | |
7M4N
 
 | |
2BIV
 
 | | Crystal structure of the wild-type MBT domains of Human SCML2 | | Descriptor: | IODIDE ION, SEX COMB ON MIDLEG-LIKE PROTEIN 2, SODIUM ION | | Authors: | Santiveri, C.M, Allen, M.D, Sait, F, Bycroft, M. | | Deposit date: | 2005-01-26 | | Release date: | 2005-02-08 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The Malignant Brain Tumor Repeats of Human Scml2 Bind to Peptides Containing Monomethylated Lysine.
J.Mol.Biol., 382, 2008
|
|
8DGN
 
 | |
8DGM
 
 | | 14-3-3 epsilon bound to phosphorylated PEAK1 (pT1165) peptide | | Descriptor: | 1,2-ETHANEDIOL, 14-3-3 protein epsilon, Inactive tyrosine-protein kinase PEAK1 | | Authors: | Roy, M.J, Hardy, J.M, Lucet, I.S. | | Deposit date: | 2022-06-24 | | Release date: | 2023-06-07 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural mapping of PEAK pseudokinase interactions identifies 14-3-3 as a molecular switch for PEAK3 signaling.
Nat Commun, 14, 2023
|
|
8DGO
 
 | |
8DGP
 
 | | 14-3-3 epsilon bound to phosphorylated PEAK3 (pS69) peptide | | Descriptor: | 1,2-ETHANEDIOL, 14-3-3 protein epsilon, Phosphorylated PEAK3 (pS69) peptide, ... | | Authors: | Roy, M.J, Hardy, J.M, Lucet, I.S. | | Deposit date: | 2022-06-24 | | Release date: | 2023-06-07 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural mapping of PEAK pseudokinase interactions identifies 14-3-3 as a molecular switch for PEAK3 signaling.
Nat Commun, 14, 2023
|
|
