5X7O
 
 | | Crystal structure of Paenibacillus sp. 598K alpha-1,6-glucosyltransferase | | Descriptor: | 1,2-ETHANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, ... | | Authors: | Fujimoto, Z, Suzuki, N, Kishine, N, Momma, M, Ichinose, H, Kimura, A, Funane, K. | | Deposit date: | 2017-02-27 | | Release date: | 2017-07-26 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Carbohydrate-binding architecture of the multi-modular alpha-1,6-glucosyltransferase from Paenibacillus sp. 598K, which produces alpha-1,6-glucosyl-alpha-glucosaccharides from starch
Biochem. J., 474, 2017
|
|
5X7G
 
 | | Crystal Structure of Paenibacillus sp. 598K cycloisomaltooligosaccharide glucanotransferase | | Descriptor: | CALCIUM ION, Cycloisomaltooligosaccharide glucanotransferase, GLYCEROL, ... | | Authors: | Fujimoto, Z, Kishine, N, Suzuki, N, Suzuki, R, Momma, M, Funane, K. | | Deposit date: | 2017-02-26 | | Release date: | 2017-04-26 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Isomaltooligosaccharide-binding structure ofPaenibacillussp. 598K cycloisomaltooligosaccharide glucanotransferase
Biosci. Rep., 37, 2017
|
|
1H88
 
 | | CRYSTAL STRUCTURE OF TERNARY PROTEIN-DNA COMPLEX1 | | Descriptor: | AMMONIUM ION, CCAAT/ENHANCER BINDING PROTEIN BETA, DNA(5'-(*CP*CP*AP*GP*TP*CP*CP*GP*TP*TP*AP* AP*GP*GP*AP*TP*TP*GP*CP*GP*CP*CP*AP*CP*AP*T)-3'), ... | | Authors: | Tahirov, T.H, Ogata, K. | | Deposit date: | 2001-01-29 | | Release date: | 2002-01-28 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Mechanism of C-Myb-C/Ebpbeta Cooperation from Separated Sites on a Promoter
Cell(Cambridge,Mass.), 108, 2002
|
|
1HJB
 
 | |
7NS0
 
 | | Bacilladnavirus capsid structure | | Descriptor: | Capsid protein VP2 | | Authors: | Munke, A, Okamoto, K. | | Deposit date: | 2021-03-05 | | Release date: | 2022-07-20 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (2.4 Å) | | Cite: | Primordial Capsid and Spooled ssDNA Genome Structures Unravel Ancestral Events of Eukaryotic Viruses.
Mbio, 13, 2022
|
|
1H8A
 
 | | CRYSTAL STRUCTURE OF TERNARY PROTEIN-DNA COMPLEX3 | | Descriptor: | CAAT/ENHANCER BINDING PROTEIN BETA, DNA(5'-(*CP*CP*AP*GP*TP*CP*CP*GP*TP*TP*AP* AP*GP*GP*AP*TP*TP*GP*CP*GP*CP*CP*AP*CP*AP*T)-3'), DNA(5'-(*GP*AP*TP*GP*TP*GP*GP*CP*GP*CP*AP* AP*TP*CP*CP*TP*TP*AP*AP*CP*GP*GP*AP*CP*TP*G)-3'), ... | | Authors: | Tahirov, T.H, Ogata, K. | | Deposit date: | 2001-01-31 | | Release date: | 2002-01-28 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | Mechanism of C-Myb-C/Ebpbeta Cooperation from Separated Sites on a Promoter
Cell(Cambridge,Mass.), 108, 2002
|
|
5WS3
 
 | | Crystal structures of human orexin 2 receptor bound to the selective antagonist EMPA determined by serial femtosecond crystallography at SACLA | | Descriptor: | N-ethyl-2-[(6-methoxypyridin-3-yl)-(2-methylphenyl)sulfonyl-amino]-N-(pyridin-3-ylmethyl)ethanamide, OLEIC ACID, Orexin receptor type 2,GlgA glycogen synthase,Orexin receptor type 2, ... | | Authors: | Suno, R, Kimura, K, Nakane, T, Yamashita, K, Wang, J, Fujiwara, T, Yamanaka, Y, Im, D, Tsujimoto, H, Sasanuma, M, Horita, S, Hirokawa, T, Nango, E, Tono, K, Kameshima, T, Hatsui, T, Joti, Y, Yabashi, M, Shimamoto, K, Yamamoto, M, Rosenbaum, D.M, Iwata, S, Shimamura, T, Kobayashi, T. | | Deposit date: | 2016-12-05 | | Release date: | 2017-12-13 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structures of Human Orexin 2 Receptor Bound to the Subtype-Selective Antagonist EMPA.
Structure, 26, 2018
|
|
7XWA
 
 | | Crystal structure of the receptor binding domain of SARS-CoV-2 Omicron BA.4/5 variant spike protein in complex with its receptor ACE2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Processed angiotensin-converting enzyme 2, ... | | Authors: | Suzuki, T, Kimura, K, Hashiguchi, T. | | Deposit date: | 2022-05-26 | | Release date: | 2022-09-28 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.36 Å) | | Cite: | Virological characteristics of the SARS-CoV-2 Omicron BA.2 subvariants, including BA.4 and BA.5.
Cell, 185, 2022
|
|
7YV1
 
 | | Human K-Ras G12D (GDP-bound) in complex with cyclic peptide inhibitor LUNA18 and KA30L Fab | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, Isoform 2B of GTPase KRas, KA30L Fab H-chain, ... | | Authors: | Irie, M, Fukami, T.A, Matsuo, A, Saka, K, Nishimura, M, Saito, H, Torizawa, T, Tanada, M, Ohta, A. | | Deposit date: | 2022-08-18 | | Release date: | 2023-07-26 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.454 Å) | | Cite: | Validation of a New Methodology to Create Oral Drugs beyond the Rule of 5 for Intracellular Tough Targets.
J.Am.Chem.Soc., 145, 2023
|
|
7YUZ
 
 | | Human K-Ras G12D (GDP-bound) in complex with cyclic peptide inhibitor AP8784 | | Descriptor: | AP8784, GUANOSINE-5'-DIPHOSPHATE, IODIDE ION, ... | | Authors: | Irie, M, Fukami, T.A, Tanada, M, Ohta, A, Torizawa, T. | | Deposit date: | 2022-08-18 | | Release date: | 2023-07-26 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.878 Å) | | Cite: | Validation of a New Methodology to Create Oral Drugs beyond the Rule of 5 for Intracellular Tough Targets.
J.Am.Chem.Soc., 145, 2023
|
|
1UCG
 
 | | Crystal structure of Ribonuclease MC1 N71T mutant | | Descriptor: | MANGANESE (II) ION, Ribonuclease MC | | Authors: | Suzuki, A, Numata, T, Yao, M, Tanaka, I, Kimura, M. | | Deposit date: | 2003-04-14 | | Release date: | 2003-04-29 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystal structures of the ribonuclease MC1 mutants N71T and N71S in complex with 5'-GMP: structural basis for alterations in substrate specificity
Biochemistry, 42, 2003
|
|
5XHF
 
 | | Crystal structure of Trastuzumab Fab fragment bearing p-azido-L-phenylalanine | | Descriptor: | polypeptide (H chain), polypeptide (L chain) | | Authors: | Kuratani, M, Yanagisawa, T, Sakamoto, K, Yokoyama, S. | | Deposit date: | 2017-04-20 | | Release date: | 2017-12-20 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.205 Å) | | Cite: | Extensive Survey of Antibody Invariant Positions for Efficient Chemical Conjugation Using Expanded Genetic Codes.
Bioconjug. Chem., 28, 2017
|
|
1IO4
 
 | |
1HJC
 
 | |
1GV5
 
 | | CRYSTAL STRUCTURE OF C-MYB R2 | | Descriptor: | MYB PROTO-ONCOGENE PROTEIN, SODIUM ION | | Authors: | Tahirov, T.H, Ogata, K. | | Deposit date: | 2002-02-06 | | Release date: | 2003-07-03 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Crystal Structure of C-Myb DNA-Binding Domain: Specific Na+ Binding and Correlation with NMR Structure
To be Published
|
|
1GVD
 
 | | CRYSTAL STRUCTURE OF C-MYB R2 V103L MUTANT | | Descriptor: | AMMONIUM ION, MYB PROTO-ONCOGENE PROTEIN, SULFATE ION | | Authors: | Tahirov, T.H, Ogata, K. | | Deposit date: | 2002-02-08 | | Release date: | 2003-07-03 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Crystal Structure of C-Myb DNA-Binding Domain: Specific Na+ Binding and Correlation with NMR Structure
To be Published
|
|
1GV2
 
 | | CRYSTAL STRUCTURE OF C-MYB R2R3 | | Descriptor: | MYB PROTO-ONCOGENE PROTEIN, SODIUM ION | | Authors: | Tahirov, T.H, Ogata, K. | | Deposit date: | 2002-02-05 | | Release date: | 2003-07-03 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Crystal Structure of C-Myb DNA-Binding Domain: Specific Na+ Binding and Correlation with NMR Structure
To be Published
|
|
1GUU
 
 | | CRYSTAL STRUCTURE OF C-MYB R1 | | Descriptor: | MYB PROTO-ONCOGENE PROTEIN, SODIUM ION | | Authors: | Tahirov, T.H, Ogata, K. | | Deposit date: | 2002-01-30 | | Release date: | 2003-06-26 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal Structure of C-Myb DNA-Binding Domain: Specific Na+ Binding and Correlation with NMR Structure
To be Published
|
|
5X7R
 
 | | Crystal structure of Paenibacillus sp. 598K alpha-1,6-glucosyltransferase complexed with isomaltohexaose | | Descriptor: | 1,2-ETHANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 4,6-dideoxy-4-{[(1S,4R,5S,6S)-4,5,6-trihydroxy-3-(hydroxymethyl)cyclohex-2-en-1-yl]amino}-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose, ... | | Authors: | Fujimoto, Z, Kishine, N, Suzuki, N, Momma, M, Ichinose, H, Kimura, A, Funane, K. | | Deposit date: | 2017-02-27 | | Release date: | 2017-07-26 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Carbohydrate-binding architecture of the multi-modular alpha-1,6-glucosyltransferase from Paenibacillus sp. 598K, which produces alpha-1,6-glucosyl-alpha-glucosaccharides from starch
Biochem. J., 474, 2017
|
|
5X7P
 
 | | Crystal structure of Paenibacillus sp. 598K alpha-1,6-glucosyltransferase complexed with acarbose | | Descriptor: | 1,2-ETHANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 4,6-dideoxy-4-{[(1S,4R,5S,6S)-4,5,6-trihydroxy-3-(hydroxymethyl)cyclohex-2-en-1-yl]amino}-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose, ... | | Authors: | Fujimoto, Z, Kishine, N, Suzuki, N, Momma, M, Ichinose, H, Kimura, A, Funane, K. | | Deposit date: | 2017-02-27 | | Release date: | 2017-07-26 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Carbohydrate-binding architecture of the multi-modular alpha-1,6-glucosyltransferase from Paenibacillus sp. 598K, which produces alpha-1,6-glucosyl-alpha-glucosaccharides from starch
Biochem. J., 474, 2017
|
|
5X7H
 
 | | Crystal Structure of Paenibacillus sp. 598K cycloisomaltooligosaccharide glucanotransferase complexed with cycloisomaltoheptaose | | Descriptor: | CALCIUM ION, Cycloisomaltooligosaccharide glucanotransferase, MALONATE ION, ... | | Authors: | Fujimoto, Z, Kishine, N, Suzuki, N, Suzuki, R, Momma, M, Funane, K. | | Deposit date: | 2017-02-26 | | Release date: | 2017-04-26 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Isomaltooligosaccharide-binding structure ofPaenibacillussp. 598K cycloisomaltooligosaccharide glucanotransferase
Biosci. Rep., 37, 2017
|
|
7EXW
 
 | | GH127 beta-L-arabinofuranosidase HypBA1 covalently complexed with alpha-L-arabinofuranosylamide | | Descriptor: | 2-bromanyl-N-[(2R,3R,4R,5S}-5-(hydroxymethyl)-3,4-bis(oxidanyl)oxolan-2-yl]ethanamide, Non-reducing end beta-L-arabinofuranosidase, ZINC ION | | Authors: | Sawano, K, Arakawa, T, Yamada, C, Fujita, K, Fushinobu, S. | | Deposit date: | 2021-05-28 | | Release date: | 2021-11-17 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Substrate complex structure, active site labeling and catalytic role of the zinc ion in cysteine glycosidase.
Glycobiology, 32, 2022
|
|
7EXV
 
 | | GH127 beta-L-arabinofuranosidase HypBA1 covalently complexed with beta-L-arabinofuranoylamide | | Descriptor: | 2-bromanyl-N-[(2S,3R,4R,5S)-5-(hydroxymethyl)-3,4-bis(oxidanyl)oxolan-2-yl]ethanamide, Non-reducing end beta-L-arabinofuranosidase, ZINC ION | | Authors: | Sawano, K, Arakawa, T, Yamada, C, Fujita, K, Fushinobu, S. | | Deposit date: | 2021-05-28 | | Release date: | 2021-11-17 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Substrate complex structure, active site labeling and catalytic role of the zinc ion in cysteine glycosidase.
Glycobiology, 32, 2022
|
|
7EXU
 
 | | GH127 beta-L-arabinofuranosidase HypBA1 E322Q mutant complexed with p-nitrophenyl beta-L-arabinofuranoside | | Descriptor: | (2S,3R,4R,5R)-2-(hydroxymethyl)-5-(4-nitrophenoxy)oxolane-3,4-diol, Non-reducing end beta-L-arabinofuranosidase, ZINC ION | | Authors: | Maruyama, S, Arakawa, T, Yamada, C, Fujita, K, Fushinobu, S. | | Deposit date: | 2021-05-28 | | Release date: | 2021-11-17 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Substrate complex structure, active site labeling and catalytic role of the zinc ion in cysteine glycosidase.
Glycobiology, 32, 2022
|
|
8JYM
 
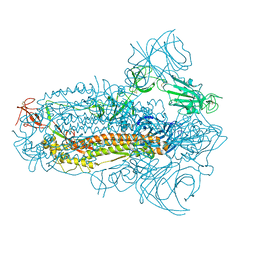 | | Structure of the SARS-CoV-2 XBB.1.5 spike glycoprotein (closed state 2) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Yajima, H, Anraku, Y, Kita, S, Kimura, K, Sasaki, J, Sasaki-Tabata, K, Maenaka, K, Hashiguchi, T. | | Deposit date: | 2023-07-03 | | Release date: | 2024-01-03 | | Method: | ELECTRON MICROSCOPY (2.79 Å) | | Cite: | Virological characteristics of the SARS-CoV-2 XBB.1.5 variant
To Be Published
|
|
