3VIU
 
 | | Crystal structure of PurL from thermus thermophilus | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, DI(HYDROXYETHYL)ETHER, MAGNESIUM ION, ... | | Authors: | Suzuki, S, Yanai, H, Kanagawa, M, Tamura, S, Watanabe, Y, Fuse, K, Baba, S, Sampei, G, Kawai, G, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2011-10-12 | | Release date: | 2012-01-18 | | Last modified: | 2012-04-25 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structure of N-formylglycinamide ribonucleotide amidotransferase II (PurL) from Thermus thermophilus HB8
Acta Crystallogr.,Sect.F, 68, 2012
|
|
2YYA
 
 | | Crystal structure of GAR synthetase from Aquifex aeolicus | | Descriptor: | Phosphoribosylamine--glycine ligase | | Authors: | Baba, S, Kanagawa, M, Kuramitsu, S, Yokoyama, S, Kawai, G, Sampei, G, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-04-27 | | Release date: | 2007-10-30 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structures of glycinamide ribonucleotide synthetase, PurD, from thermophilic eubacteria
J.Biochem., 148, 2010
|
|
2YW2
 
 | | Crystal structure of GAR synthetase from Aquifex aeolicus in complex with ATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, PHOSPHATE ION, Phosphoribosylamine--glycine ligase | | Authors: | Baba, S, Kanagawa, M, Kuramitsu, S, Yokoyama, S, Kawai, G, Sampei, G, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-04-19 | | Release date: | 2007-10-23 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structures of glycinamide ribonucleotide synthetase, PurD, from thermophilic eubacteria
J.Biochem., 148, 2010
|
|
2YRX
 
 | | Crystal structure of GAR synthetase from Geobacillus kaustophilus | | Descriptor: | ADENOSINE MONOPHOSPHATE, PHOSPHATE ION, Phosphoribosylglycinamide synthetase | | Authors: | Baba, S, Kanagawa, M, Kuramitsu, S, Yokoyama, S, Kawai, G, Sampei, G, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-04-03 | | Release date: | 2007-10-09 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structures of glycinamide ribonucleotide synthetase, PurD, from thermophilic eubacteria
J.Biochem., 148, 2010
|
|
5AVM
 
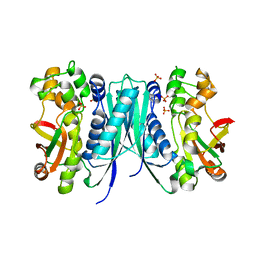 | | Crystal structures of 5-aminoimidazole ribonucleotide (AIR) synthetase, PurM, from Thermus thermophilus | | Descriptor: | Phosphoribosylformylglycinamidine cyclo-ligase, SULFATE ION | | Authors: | Kanagawa, M, Baba, S, Watanabe, Y, Nakagawa, N, Ebihara, A, Sampei, G, Kawai, G, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2015-06-23 | | Release date: | 2015-11-25 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structures and ligand binding of PurM proteins from Thermus thermophilus and Geobacillus kaustophilus
J.Biochem., 159, 2016
|
|
2YRW
 
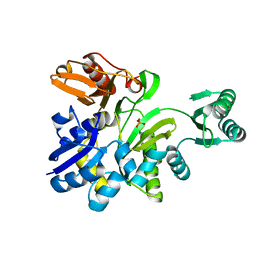 | | Crystal structure of GAR synthetase from Geobacillus kaustophilus | | Descriptor: | PHOSPHATE ION, Phosphoribosylglycinamide synthetase | | Authors: | Baba, S, Kanagawa, M, Kuramitsu, S, Yokoyama, S, Kawai, G, Sampei, G, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-04-03 | | Release date: | 2007-10-09 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structures of glycinamide ribonucleotide synthetase, PurD, from thermophilic eubacteria
J.Biochem., 148, 2010
|
|
2YS6
 
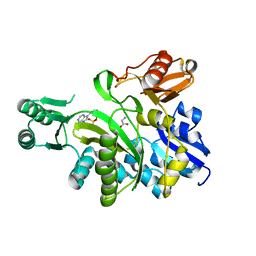 | | Crystal structure of GAR synthetase from Geobacillus kaustophilus | | Descriptor: | ADENOSINE MONOPHOSPHATE, GLYCINE, Phosphoribosylglycinamide synthetase | | Authors: | Baba, S, Kanagawa, M, Kuramitsu, S, Yokoyama, S, Kawai, G, Sampei, G, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-04-03 | | Release date: | 2007-10-09 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Crystal structures of glycinamide ribonucleotide synthetase, PurD, from thermophilic eubacteria
J.Biochem., 148, 2010
|
|
2YS7
 
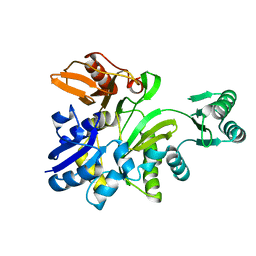 | | Crystal structure of GAR synthetase from Geobacillus kaustophilus | | Descriptor: | Phosphoribosylglycinamide synthetase | | Authors: | Baba, S, Kanagawa, M, Kuramitsu, S, Yokoyama, S, Kawai, G, Sampei, G, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-04-03 | | Release date: | 2007-10-09 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Crystal structures of glycinamide ribonucleotide synthetase, PurD, from thermophilic eubacteria
J.Biochem., 148, 2010
|
|
3AV3
 
 | | Crystal structure of glycinamide ribonucleotide transformylase 1 from Geobacillus kaustophilus | | Descriptor: | MAGNESIUM ION, Phosphoribosylglycinamide formyltransferase | | Authors: | Kanagawa, M, Baba, S, Nakagawa, N, Ebihara, A, Kuramitsu, S, Yokoyama, S, Sampei, G, Kawai, G, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2011-02-18 | | Release date: | 2012-03-07 | | Last modified: | 2014-01-15 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structures and reaction mechanisms of the two related enzymes, PurN and PurU.
J.Biochem., 154, 2013
|
|
3AUF
 
 | | Crystal structure of glycinamide ribonucleotide transformylase 1 from Symbiobacterium toebii | | Descriptor: | Glycinamide ribonucleotide transformylase 1 | | Authors: | Kanagawa, M, Baba, S, Nagira, T, Kuramitsu, S, Yokoyama, S, Sampei, G, Kawai, G, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2011-02-03 | | Release date: | 2012-03-07 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Structures and reaction mechanisms of the two related enzymes, PurN and PurU.
J.Biochem., 154, 2013
|
|
2YWR
 
 | | Crystal structure of GAR transformylase from Aquifex aeolicus | | Descriptor: | COBALT (II) ION, MAGNESIUM ION, Phosphoribosylglycinamide formyltransferase | | Authors: | Kanagawa, M, Baba, S, Kuramitsu, S, Yokoyama, S, Kawai, G, Sampei, G, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-04-23 | | Release date: | 2007-10-23 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Structures and reaction mechanisms of the two related enzymes, PurN and PurU.
J.Biochem., 154, 2013
|
|
1WKS
 
 | |
1XWP
 
 | | Solution structure of AUCGCA loop | | Descriptor: | 5'-R(*GP*GP*AP*GP*AP*UP*CP*GP*CP*AP*CP*UP*CP*CP*A)-3' | | Authors: | Sakamoto, T, Oguro, A, Kawai, G, Ohtsu, T, Nakamura, Y. | | Deposit date: | 2004-11-02 | | Release date: | 2005-02-15 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | NMR structures of double loops of an RNA aptamer against mammalian initiation factor 4A
Nucleic Acids Res., 33, 2005
|
|
1XWU
 
 | | Solution structure of ACAUAGA loop | | Descriptor: | 5'-R(*CP*GP*AP*AP*AP*CP*AP*UP*AP*GP*AP*UP*UP*CP*GP*A)-3' | | Authors: | Sakamoto, T, Oguro, A, Kawai, G, Ohtsu, T, Nakamura, Y. | | Deposit date: | 2004-11-02 | | Release date: | 2005-02-15 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | NMR structures of double loops of an RNA aptamer against mammalian initiation factor 4A
Nucleic Acids Res., 33, 2005
|
|
6IZP
 
 | | Solution structure of the complex of naphthyridine carbamate dimer and an RNA with UGGAA-UGGAA pentad | | Descriptor: | 3-[3-[(7-methyl-1,8-naphthyridin-2-yl)carbamoyloxy]propylamino]propyl ~{N}-(7-methyl-1,8-naphthyridin-2-yl)carbamate, RNA (29-MER) | | Authors: | Nagano, K, Shibata, T, Nakatani, K, Kawai, G. | | Deposit date: | 2018-12-20 | | Release date: | 2019-12-25 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Small molecule targeting r(UGGAA)n disrupts RNA foci and alleviates disease phenotype in Drosophila model
Nat Commun, 12, 2021
|
|
2D17
 
 | | Solution RNA structure of stem-bulge-stem region of the HIV-1 dimerization initiation site | | Descriptor: | 5'-R(*CP*GP*GP*CP*AP*AP*GP*AP*GP*GP*CP*GP*AP*CP*CP*C)-3', 5'-R(*GP*GP*GP*UP*CP*GP*GP*CP*UP*UP*GP*CP*UP*G)-3' | | Authors: | Baba, S, Takahashi, K, Noguchi, S, Takaku, H, Koyanagi, Y, Yamamoto, N, Kawai, G. | | Deposit date: | 2005-08-15 | | Release date: | 2005-11-01 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution RNA structures of the HIV-1 dimerization initiation site in the kissing-loop and extended-duplex dimers.
J.Biochem.(Tokyo), 138, 2005
|
|
2D1A
 
 | | Solution RNA structure model of the HIV-1 dimerization initiation site in the extended-duplex dimer | | Descriptor: | RNA | | Authors: | Baba, S, Takahashi, K, Noguchi, S, Takaku, H, Koyanagi, Y, Yamamoto, N, Kawai, G. | | Deposit date: | 2005-08-15 | | Release date: | 2005-11-01 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution RNA structures of the HIV-1 dimerization initiation site in the kissing-loop and extended-duplex dimers.
J.Biochem.(Tokyo), 138, 2005
|
|
2D18
 
 | | Solution RNA structure of loop region of the HIV-1 dimerization initiation site in the extended-duplex dimer | | Descriptor: | 5'-R(*GP*CP*UP*GP*AP*AP*GP*UP*GP*CP*AP*CP*AP*CP*GP*GP*C)-3' | | Authors: | Baba, S, Takahashi, K, Noguchi, S, Takaku, H, Koyanagi, Y, Yamamoto, N, Kawai, G. | | Deposit date: | 2005-08-15 | | Release date: | 2005-11-01 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution RNA structures of the HIV-1 dimerization initiation site in the kissing-loop and extended-duplex dimers.
J.Biochem.(Tokyo), 138, 2005
|
|
2D1B
 
 | | Solution RNA structure model of the HIV-1 dimerization initiation site in the kissing-loop dimer | | Descriptor: | RNA | | Authors: | Baba, S, Takahashi, K, Noguchi, S, Takaku, H, Koyanagi, Y, Yamamoto, N, Kawai, G. | | Deposit date: | 2005-08-15 | | Release date: | 2005-11-01 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution RNA structures of the HIV-1 dimerization initiation site in the kissing-loop and extended-duplex dimers.
J.Biochem.(Tokyo), 138, 2005
|
|
2D19
 
 | | Solution RNA structure of loop region of the HIV-1 dimerization initiation site in the kissing-loop dimer | | Descriptor: | 5'-R(*GP*CP*UP*GP*AP*AP*GP*UP*GP*CP*AP*CP*AP*CP*GP*GP*C)-3' | | Authors: | Baba, S, Takahashi, K, Noguchi, S, Takaku, H, Koyanagi, Y, Yamamoto, N, Kawai, G. | | Deposit date: | 2005-08-15 | | Release date: | 2005-11-01 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution RNA structures of the HIV-1 dimerization initiation site in the kissing-loop and extended-duplex dimers.
J.Biochem.(Tokyo), 138, 2005
|
|
2YZL
 
 | | Crystal structure of phosphoribosylaminoimidazole-succinocarboxamide synthase with ADP from Methanocaldococcus jannaschii | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Phosphoribosylaminoimidazole-succinocarboxamide synthase, SULFATE ION | | Authors: | Kanagawa, M, Baba, S, Fukui, K, Kuramitsu, S, Yokoyama, S, Kawai, G, Sampei, G, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-05-06 | | Release date: | 2007-11-06 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of phosphoribosylaminoimidazole-succinocarboxamide synthase with ADP from Methanocaldococcus jannaschii
To be Published
|
|
2YWX
 
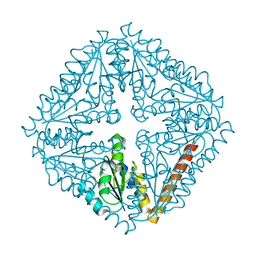 | | Crystal structure of phosphoribosylaminoimidazole carboxylase catalytic subunit from Methanocaldococcus jannaschii | | Descriptor: | Phosphoribosylaminoimidazole carboxylase catalytic subunit | | Authors: | Kanagawa, M, Baba, S, Kuramitsu, S, Yokoyama, S, Kawai, G, Sampei, G, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-04-23 | | Release date: | 2007-10-23 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Crystal structure of phosphoribosylaminoimidazole carboxylase catalytic subunit from Methanocaldococcus jannaschii
To be Published
|
|
2YX5
 
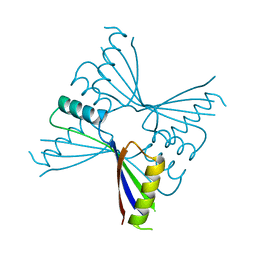 | | Crystal Structure of Methanocaldococcus jannaschii PurS, One of the Subunits of Formylglycinamide Ribonucleotide Amidotransferase in the Purine Biosynthetic Pathway | | Descriptor: | UPF0062 protein MJ1593 | | Authors: | Kanagawa, M, Baba, S, Agari, Y, Chen, L.Q, Fu, Z.-Q, Chrzas, J, Wang, B.C, Kuramitsu, S, Yokoyama, S, Kawai, G, Sampei, G, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-04-24 | | Release date: | 2007-10-30 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structure of Methanocaldococcus jannaschii PurS, One of the Subunits of Formylglycinamide Ribonucleotide Amidotransferase in the Purine Biosynthetic Pathway
To be Published
|
|
2YWV
 
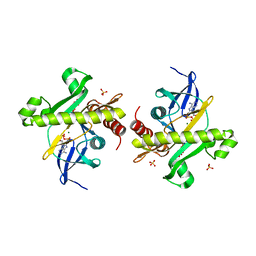 | | Crystal structure of SAICAR synthetase from Geobacillus kaustophilus | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, Phosphoribosylaminoimidazole succinocarboxamide synthetase, ... | | Authors: | Kanagawa, M, Baba, S, Kuramitsu, S, Yokoyama, S, Kawai, G, Sampei, G, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-04-23 | | Release date: | 2007-10-23 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal structure of SAICAR synthetase from Geobacillus kaustophilus
To be Published
|
|
2YWB
 
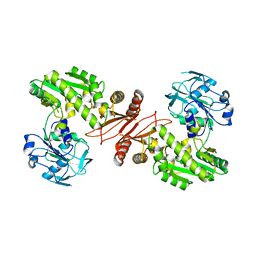 | | Crystal structure of GMP synthetase from Thermus thermophilus | | Descriptor: | GMP synthase [glutamine-hydrolyzing] | | Authors: | Baba, S, Kanagawa, M, Yanai, H, Ishii, T, Kuramitsu, S, Yokoyama, S, Sampei, G, Kawai, G, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-04-20 | | Release date: | 2007-10-23 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of GMP synthetase from Thermus thermophilus
To be Published
|
|
