8A4L
 
 | | Lipidic alpha-synuclein fibril - polymorph L2A | | Descriptor: | Alpha-synuclein | | Authors: | Frieg, B, Antonschmidt, L, Dienemann, C, Geraets, J.A, Najbauer, E.E, Matthes, D, de Groot, B.L, Andreas, L.B, Becker, S, Griesinger, C, Schroeder, G.F. | | Deposit date: | 2022-06-12 | | Release date: | 2022-08-17 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (2.68 Å) | | Cite: | The 3D structure of lipidic fibrils of alpha-synuclein.
Nat Commun, 13, 2022
|
|
6GEZ
 
 | | THE STRUCTURE OF TWITCH-2B N532F | | Descriptor: | CALCIUM ION, FORMIC ACID, Green fluorescent protein,Optimized Ratiometric Calcium Sensor,Green fluorescent protein,Green fluorescent protein | | Authors: | Trigo Mourino, P, Paulat, M, Thestrup, T, Griesbeck, O, Griesinger, C, Becker, S. | | Deposit date: | 2018-04-27 | | Release date: | 2019-08-21 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.47 Å) | | Cite: | Dynamic tuning of FRET in a green fluorescent protein biosensor.
Sci Adv, 5, 2019
|
|
2Y0R
 
 | | Structural basis for the allosteric interference of myosin function by mutants G680A and G680V of Dictyostelium myosin-2 | | Descriptor: | MYOSIN-2 HEAVY CHAIN | | Authors: | Preller, M, Bauer, S, Adamek, N, Fujita-Becker, S, Fedorov, R, Geeves, M.A, Manstein, D.J. | | Deposit date: | 2010-12-07 | | Release date: | 2011-07-20 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Structural Basis for the Allosteric Interference of Myosin Function by Reactive Thiol Region Mutations G680A and G680V.
J.Biol.Chem., 286, 2011
|
|
2Y8I
 
 | | Structural basis for the allosteric interference of myosin function by mutants G680A and G680V of Dictyostelium myosin-2 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, MYOSIN-2 HEAVY CHAIN | | Authors: | Preller, M, Bauer, S, Adamek, N, Fujita-Becker, S, Fedorov, R, Geeves, M.A, Manstein, D.J. | | Deposit date: | 2011-02-07 | | Release date: | 2011-07-20 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.132 Å) | | Cite: | Structural Basis for the Allosteric Interference of Myosin Function by Reactive Thiol Region Mutations G680A and G680V.
J.Biol.Chem., 286, 2011
|
|
5NWX
 
 | | Insight into the molecular recognition mechanism of the coactivator NCoA1 by STAT6 | | Descriptor: | Nuclear receptor coactivator 1, Signal transducer and activator of transcription 6 | | Authors: | Russo, L, Giller, K, Pfitzner, E, Griesinger, C, Becker, S. | | Deposit date: | 2017-05-08 | | Release date: | 2017-12-13 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | Insight into the molecular recognition mechanism of the coactivator NCoA1 by STAT6.
Sci Rep, 7, 2017
|
|
6GEL
 
 | | The structure of TWITCH-2B | | Descriptor: | CALCIUM ION, FORMIC ACID, GLYCEROL, ... | | Authors: | Trigo Mourino, P, Paulat, M, Thestrup, T, Griesbeck, O, Griesinger, C, Becker, S. | | Deposit date: | 2018-04-26 | | Release date: | 2019-08-21 | | Last modified: | 2019-09-11 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | Dynamic tuning of FRET in a green fluorescent protein biosensor.
Sci Adv, 5, 2019
|
|
2JK4
 
 | | Structure of the human voltage-dependent anion channel | | Descriptor: | VOLTAGE-DEPENDENT ANION-SELECTIVE CHANNEL PROTEIN 1 | | Authors: | Bayrhuber, M, Meins, T, Habeck, M, Becker, S, Giller, K, Villinger, S, Vonrhein, C, Griesinger, C, Zweckstetter, M, Zeth, K. | | Deposit date: | 2008-08-15 | | Release date: | 2008-10-14 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (4.1 Å) | | Cite: | Structure of the Human Voltage-Dependent Anion Channel.
Proc.Natl.Acad.Sci.USA, 105, 2008
|
|
5JDP
 
 | | E73V mutant of the human voltage-dependent anion channel | | Descriptor: | Voltage-dependent anion-selective channel protein 1 | | Authors: | Jaremko, M, Jaremko, L, Villinger, S, Schmidt, C, Giller, K, Griesinger, C, Becker, S, Zweckstetter, M. | | Deposit date: | 2016-04-17 | | Release date: | 2016-08-10 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | High-Resolution NMR Determination of the Dynamic Structure of Membrane Proteins.
Angew.Chem.Int.Ed.Engl., 55, 2016
|
|
8P7K
 
 | | The impact of molecular variants, crystallization conditions and space group on structure-ligand complexes: A case study on Bacterial Phosphotriesterase Variants and complexes | | Descriptor: | (2~{R})-2-methylpentanedioic acid, FORMIC ACID, Parathion hydrolase, ... | | Authors: | Dym, O, Aggawal, N, Ashani, Y, Albeck, S, Unger, T, Hamer Rogotner, S, Silman, I, Sussman, J.L. | | Deposit date: | 2023-05-30 | | Release date: | 2023-06-07 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.929 Å) | | Cite: | The impact of molecular variants, crystallization conditions and the space group on ligand-protein complexes: a case study on bacterial phosphotriesterase.
Acta Crystallogr D Struct Biol, 79, 2023
|
|
1G8X
 
 | | STRUCTURE OF A GENETICALLY ENGINEERED MOLECULAR MOTOR | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, MYOSIN II HEAVY CHAIN FUSED TO ALPHA-ACTININ 3 | | Authors: | Kliche, W, Fujita-Becker, S, Kollmar, M, Manstein, D.J, Kull, F.J. | | Deposit date: | 2000-11-21 | | Release date: | 2001-01-17 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of a genetically engineered molecular motor.
EMBO J., 20, 2001
|
|
5FQ1
 
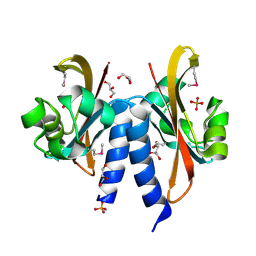 | |
7O4D
 
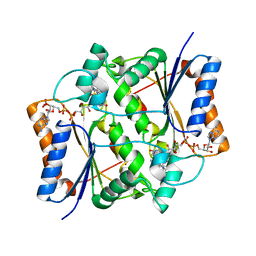 | | QR2 inhibitor from a novel sulfanamide series to tackle age related oxidative stress and cognitive decline | | Descriptor: | 8-methyl-2-(4-methyl-3-piperazin-1-ylsulfonyl-phenyl)imidazo[1,2-a]pyridine, FLAVIN-ADENINE DINUCLEOTIDE, Ribosyldihydronicotinamide dehydrogenase [quinone], ... | | Authors: | Gould, N.L, Scherer, G.R, Carvalh, S, Shurrush, K, Edry, E, Elkobi, A, David, O, Dym, O, Albeck, S, Peleg, Y, Germain, N, Babaev, I, Sharir, H, Lefker, B, Subramanyam, C, Barr, H, Rosenblum, K. | | Deposit date: | 2021-04-06 | | Release date: | 2022-08-17 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.249 Å) | | Cite: | Specific quinone reductase 2 inhibitors reduce metabolic burden and reverse Alzheimer's disease phenotype in mice.
J.Clin.Invest., 133, 2023
|
|
5ABS
 
 | | CRYSTAL STRUCTURE OF THE C-TERMINAL COILED-COIL DOMAIN OF CIN85 IN SPACE GROUP P321 | | Descriptor: | SH3 DOMAIN-CONTAINING KINASE-BINDING PROTEIN 1, ZINC ION | | Authors: | Wong, L, Habeck, M, Griesinger, C, Becker, S. | | Deposit date: | 2015-08-07 | | Release date: | 2016-07-13 | | Last modified: | 2019-02-06 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | The Adaptor Protein Cin85 Assembles Intracellular Signaling Clusters for B Cell Activation.
Sci.Signal., 9, 2016
|
|
1Y1U
 
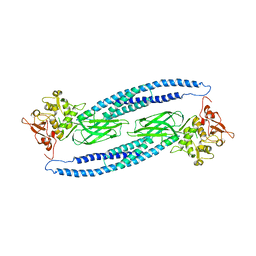 | | Structure of unphosphorylated STAT5a | | Descriptor: | Signal transducer and activator of transcription 5A | | Authors: | Neculai, D, Neculai, A.M, Verrier, S, Straub, K, Klumpp, K, Pfitzner, E, Becker, S. | | Deposit date: | 2004-11-19 | | Release date: | 2005-10-04 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3.21 Å) | | Cite: | Structure of the unphosphorylated STAT5a dimer
J.Biol.Chem., 280, 2005
|
|
2V8Z
 
 | | Crystal Structure of YagE, a prophage protein belonging to the dihydrodipicolinic acid synthase family from E. coli K12 | | Descriptor: | YAGE | | Authors: | Manicka, S, Peleg, Y, Unger, T, Albeck, S, Dym, O, Greenblatt, H.M, Bourenkov, G, Lamzin, V, Krishnaswamy, S, Sussman, J.L. | | Deposit date: | 2007-08-16 | | Release date: | 2008-03-04 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal Structure of Yage, a Putative Dhdps Like Protein from Escherichia Coli K12.
Proteins: Struct., Funct., Bioinf., 71, 2008
|
|
1UTX
 
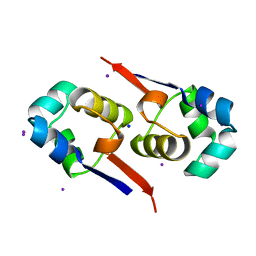 | | Regulation of Cytolysin Expression by Enterococcus faecalis: Role of CylR2 | | Descriptor: | CYLR2, IODIDE ION, SODIUM ION | | Authors: | Razeto, A, Rumpel, S, Pillar, C.M, Gilmore, M.S, Becker, S, Zweckstetter, M. | | Deposit date: | 2003-12-12 | | Release date: | 2004-09-16 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure and DNA-Binding Properties of the Cytolysin Regulator CylR2 from Enterococcus Faecalis
Embo J., 23, 2004
|
|
2V9D
 
 | | Crystal Structure of YagE, a prophage protein belonging to the dihydrodipicolinic acid synthase family from E. coli K12 | | Descriptor: | YAGE | | Authors: | Manicka, S, Peleg, Y, Unger, T, Albeck, S, Dym, O, Greenblatt, H.M, Bourenkov, G, Lamzin, V, Krishnaswamy, S, Sussman, J.L. | | Deposit date: | 2007-08-23 | | Release date: | 2008-03-04 | | Last modified: | 2019-11-06 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Crystal Structure of Yage, a Putative Dhdps Like Protein from Escherichia Coli K12.
Proteins: Struct., Funct., Bioinf., 71, 2008
|
|
3GWH
 
 | | Crystallographic Ab Initio protein solution far below atomic resolution | | Descriptor: | PHOSPHATE ION, Transcriptional antiterminator (BglG family) | | Authors: | Rodriguez, D.D, Grosse, C, Himmel, S, Gonzalez, C, Becker, S, Sheldrick, G.M, Uson, I. | | Deposit date: | 2009-04-01 | | Release date: | 2010-04-07 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystallographic ab initio protein structure solution below atomic resolution
Nat.Methods, 6, 2009
|
|
2N64
 
 | |
2YAT
 
 | | Crystal structure of estradiol derived metal chelate and estrogen receptor-ligand binding domain complex | | Descriptor: | ESTRADIOL-PYRIDINIUM TETRAACETIC ACID, ESTROGEN RECEPTOR, EUROPIUM ION, ... | | Authors: | Li, M.J, Greenblatt, H.M, Dym, O, Albeck, S, Degani, H, Sussman, J.L. | | Deposit date: | 2011-02-24 | | Release date: | 2011-06-01 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.602 Å) | | Cite: | Structure of Estradiol Metal Chelate and Estrogen Receptor Complex: The Basis for Designing a New Class of Selective Estrogen Receptor Modulators.
J.Med.Chem., 54, 2011
|
|
3BTP
 
 | | Crystal structure of Agrobacterium tumefaciens VirE2 in complex with its chaperone VirE1: a novel fold and implications for DNA binding | | Descriptor: | AMMONIUM ION, DI(HYDROXYETHYL)ETHER, Protein virE1, ... | | Authors: | Dym, O, Albeck, S, Unger, T, Elbaum, M, Israel Structural Proteomics Center (ISPC) | | Deposit date: | 2007-12-30 | | Release date: | 2008-08-19 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of the Agrobacterium virulence complex VirE1-VirE2 reveals a flexible protein that can accommodate different partners.
Proc.Natl.Acad.Sci.Usa, 105, 2008
|
|
8P7I
 
 | | The impact of molecular variants, crystallization conditions and space group on structure-ligand complexes: A case study on Bacterial Phosphotriesterase Variants and complexes | | Descriptor: | (2~{S},6~{R})-2,6-dimethyl-1,3-dioxane-4,4-diol, FORMIC ACID, Parathion hydrolase, ... | | Authors: | Dym, O, Aggarwal, N, Ashani, Y, Albeck, S, Unger, T, Hamer Rogotner, S, Silman, I, Sussman, J.L. | | Deposit date: | 2023-05-30 | | Release date: | 2023-10-25 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The impact of molecular variants, crystallization conditions and the space group on ligand-protein complexes: a case study on bacterial phosphotriesterase.
Acta Crystallogr D Struct Biol, 79, 2023
|
|
8P7N
 
 | | The impact of molecular variants, crystallization conditions and space group on structure-ligand complexes: A case study on Bacterial Phosphotriesterase Variants and complexes | | Descriptor: | FORMIC ACID, Parathion hydrolase, ZINC ION | | Authors: | Dym, O, Aggarwal, N, Ashani, Y, Albeck, S, Unger, T, Hamer Rogotner, S, Silman, I, Sussman, J.L. | | Deposit date: | 2023-05-30 | | Release date: | 2023-10-25 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | The impact of molecular variants, crystallization conditions and the space group on ligand-protein complexes: a case study on bacterial phosphotriesterase.
Acta Crystallogr D Struct Biol, 79, 2023
|
|
8P7T
 
 | | The impact of molecular variants, crystallization conditions and space group on structure-ligand complexes: A case study on Bacterial Phosphotriesterase Variants and complexes | | Descriptor: | 1-ethyl-1-methyl-cyclohexane, FORMIC ACID, GLYCEROL, ... | | Authors: | Dym, O, Aggarwal, N, Ashani, Y, Albeck, S, Unger, T, Hamer Rogotner, S, Silman, I, Sussman, J.L. | | Deposit date: | 2023-05-31 | | Release date: | 2023-10-25 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The impact of molecular variants, crystallization conditions and the space group on ligand-protein complexes: a case study on bacterial phosphotriesterase.
Acta Crystallogr D Struct Biol, 79, 2023
|
|
8P7V
 
 | | The impact of molecular variants, crystallization conditions and space group on structure-ligand complexes: A case study on Bacterial Phosphotriesterase Variants and complexes | | Descriptor: | 1,2-ETHANEDIOL, 1-ethyl-1-methyl-cyclohexane, FORMIC ACID, ... | | Authors: | Dym, O, Aggawal, N, Ashani, Y, Albeck, S, Unger, T, Hamer Rogotner, S, Silman, I, Sussman, J.L. | | Deposit date: | 2023-05-31 | | Release date: | 2023-10-25 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.737 Å) | | Cite: | The impact of molecular variants, crystallization conditions and the space group on ligand-protein complexes: a case study on bacterial phosphotriesterase.
Acta Crystallogr D Struct Biol, 79, 2023
|
|
