6WFN
 
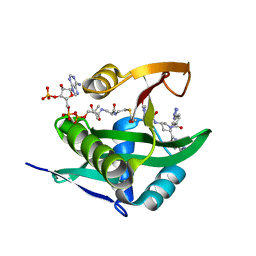 | | Crystal structure of human Naa50 in complex with AcCoA and an inhibitor (compound 4a) identified using DNA encoded library technology | | Descriptor: | (4S)-1-methyl-N-{(3S,5S)-5-[4-(methylcarbamoyl)-1,3-thiazol-2-yl]-1-[4-(1H-tetrazol-5-yl)benzene-1-carbonyl]pyrrolidin-3-yl}-2,6-dioxohexahydropyrimidine-4-carboxamide, ACETYL COENZYME *A, N-alpha-acetyltransferase 50 | | Authors: | Greasley, S.E, Feng, J, Deng, Y.-L, Stewart, A.E. | | Deposit date: | 2020-04-03 | | Release date: | 2020-07-01 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.07 Å) | | Cite: | Characterization of SpecificN-alpha-Acetyltransferase 50 (Naa50) Inhibitors Identified Using a DNA Encoded Library.
Acs Med.Chem.Lett., 11, 2020
|
|
6WFK
 
 | | Crystal structure of human Naa50 in complex with CoA and an inhibitor (compound 4a) identified using DNA encoded library technology | | Descriptor: | (4S)-1-methyl-N-{(3S,5S)-5-[4-(methylcarbamoyl)-1,3-thiazol-2-yl]-1-[4-(1H-tetrazol-5-yl)benzene-1-carbonyl]pyrrolidin-3-yl}-2,6-dioxohexahydropyrimidine-4-carboxamide, COENZYME A, N-alpha-acetyltransferase 50 | | Authors: | Greasley, S.E, Feng, J, Deng, Y.-L, Stewart, A.E. | | Deposit date: | 2020-04-03 | | Release date: | 2020-07-01 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Characterization of SpecificN-alpha-Acetyltransferase 50 (Naa50) Inhibitors Identified Using a DNA Encoded Library.
Acs Med.Chem.Lett., 11, 2020
|
|
6LS5
 
 | | Structure of human liver FBPase complexed with covalent allosteric inhibitor | | Descriptor: | 2-(ethyldisulfanyl)-1,3-benzothiazole, ADENOSINE MONOPHOSPHATE, Fructose-1,6-bisphosphatase 1, ... | | Authors: | Yunyuan, H, Rongrong, S, Yixiang, X, Shuaishuai, N, Yanliang, R, Jian, L, Jian, W. | | Deposit date: | 2020-01-17 | | Release date: | 2020-05-27 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.031 Å) | | Cite: | Identification of the New Covalent Allosteric Binding Site of Fructose-1,6-bisphosphatase with Disulfiram Derivatives toward Glucose Reduction.
J.Med.Chem., 63, 2020
|
|
7LJX
 
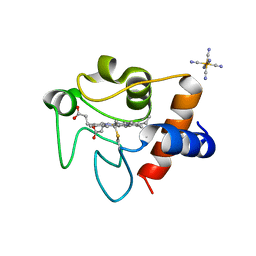 | | Oxidized rat cytochrome c mutant (K53Q) | | Descriptor: | Cytochrome c, somatic, HEME C, ... | | Authors: | Huttemann, M, Edwards, B.F.P, Brunzelle, J.S, Vaishnav, A. | | Deposit date: | 2021-02-01 | | Release date: | 2021-05-12 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.31 Å) | | Cite: | Lysine 53 Acetylation of Cytochrome c in Prostate Cancer: Warburg Metabolism and Evasion of Apoptosis.
Cells, 10, 2021
|
|
5TYT
 
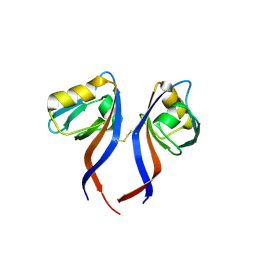 | | Crystal Structure of the PDZ domain of RhoGEF bound to CXCR2 C-terminal peptide | | Descriptor: | Rho guanine nucleotide exchange factor 11, C-X-C chemokine receptor type 2 chimera | | Authors: | Spellmon, N, Holcomb, J, Niu, A, Choudhary, V, Sun, X, Brunzelle, J, Li, C, Yang, Z. | | Deposit date: | 2016-11-21 | | Release date: | 2017-02-22 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.398 Å) | | Cite: | Structural basis of PDZ-mediated chemokine receptor CXCR2 scaffolding by guanine nucleotide exchange factor PDZ-RhoGEF.
Biochem. Biophys. Res. Commun., 485, 2017
|
|
5ET0
 
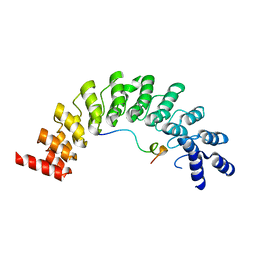 | | Crystal structure of Myo3b-ARB2 in complex with Espin1-AR | | Descriptor: | Espin, Myosin-IIIb | | Authors: | Liu, H, Li, J, Liu, W, Zhang, M. | | Deposit date: | 2015-11-17 | | Release date: | 2016-02-03 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Myosin III-mediated cross-linking and stimulation of actin bundling activity of Espin
Elife, 5, 2016
|
|
5ET1
 
 | | Crystal structure of Myo3b-ARB1 in complex with Espin1-AR | | Descriptor: | Espin, GLYCEROL, Myosin-IIIb | | Authors: | Liu, H, Li, J, liu, W, Zhang, M. | | Deposit date: | 2015-11-17 | | Release date: | 2016-02-03 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Myosin III-mediated cross-linking and stimulation of actin bundling activity of Espin
Elife, 5, 2016
|
|
4NLD
 
 | | Crystal structure of the hepatitis C virus NS5B RNA-dependent RNA polymerase complex with BMS-791325 also known as (1aR,12bS)-8-cyclohexyl-n-(dimethylsulfamoyl)-11-methoxy-1a-{[(1R,5S)-3-methyl-3,8-diazabicyclo[3.2.1]oct-8-yl]carbonyl}-1,1a,2,12b-tetrahydrocyclopropa[d]indolo[2,1-a][2]benzazepine-5-carboxamide and 2-(4-fluorophenyl)-n-methyl-6-[(methylsulfonyl)amino]-5-(propan-2-yloxy)-1-benzofuran-3-carboxamide | | Descriptor: | (1aR,12bS)-8-cyclohexyl-N-(dimethylsulfamoyl)-11-methoxy-1a-{[(1R,5S)-3-methyl-3,8-diazabicyclo[3.2.1]oct-8-yl]carbonyl}-1,1a,2,12b-tetrahydrocyclopropa[d]indolo[2,1-a][2]benzazepine-5-carboxamide, 2-(4-fluorophenyl)-N-methyl-6-[(methylsulfonyl)amino]-5-(propan-2-yloxy)-1-benzofuran-3-carboxamide, RNA-directed RNA polymerase, ... | | Authors: | Sheriff, S. | | Deposit date: | 2013-11-14 | | Release date: | 2014-03-19 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Discovery and Preclinical Characterization of the Cyclopropylindolobenzazepine BMS-791325, A Potent Allosteric Inhibitor of the Hepatitis C Virus NS5B Polymerase.
J.Med.Chem., 57, 2014
|
|
5VB3
 
 | | X-ray structure of nuclear receptor ROR-gammat Ligand Binding Domain + SRC2 peptide | | Descriptor: | Nuclear receptor ROR-gamma, SRC2 chimera, SODIUM ION | | Authors: | Li, X. | | Deposit date: | 2017-03-28 | | Release date: | 2017-06-07 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural studies unravel the active conformation of apo ROR gamma t nuclear receptor and a common inverse agonism of two diverse classes of ROR gamma t inhibitors.
J. Biol. Chem., 292, 2017
|
|
5VB7
 
 | | X-ray co-structure of nuclear receptor ROR-gammat Ligand Binding Domain with an agonist and SRC2 peptide | | Descriptor: | N-methyl-N'-(3-methylbut-2-en-1-yl)-N'-(3-phenoxyphenyl)-N-[trans-4-(pyridin-4-yl)cyclohexyl]urea, Nuclear receptor ROR-gamma, SRC2 chimera, ... | | Authors: | Li, X. | | Deposit date: | 2017-03-28 | | Release date: | 2017-06-07 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.335 Å) | | Cite: | Structural studies unravel the active conformation of apo ROR gamma t nuclear receptor and a common inverse agonism of two diverse classes of ROR gamma t inhibitors.
J. Biol. Chem., 292, 2017
|
|
5VB6
 
 | |
5VB5
 
 | | X-ray co-structure of nuclear receptor ROR-gammat Ligand Binding Domain with an inverse agonist and SRC2 peptide | | Descriptor: | N-[(2R)-3-(4-{[3-(4-chlorophenyl)propanoyl]amino}phenyl)-1-(4-methylpiperidin-1-yl)-1-oxopropan-2-yl]-4-methylpentanamide, Nuclear receptor ROR-gamma, SRC2 chimera, ... | | Authors: | Li, X. | | Deposit date: | 2017-03-28 | | Release date: | 2017-06-07 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.226 Å) | | Cite: | Structural studies unravel the active conformation of apo ROR gamma t nuclear receptor and a common inverse agonism of two diverse classes of ROR gamma t inhibitors.
J. Biol. Chem., 292, 2017
|
|
5ZZ9
 
 | | Crystal structure of Homer2 EVH1/Drebrin PPXXF complex | | Descriptor: | Homer protein homolog 2, Peptide from Drebrin | | Authors: | Li, Z, Liu, H, Li, J, Liu, W, Zhang, M. | | Deposit date: | 2018-05-31 | | Release date: | 2018-12-19 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Homer Tetramer Promotes Actin Bundling Activity of Drebrin.
Structure, 27, 2019
|
|
3Q0Z
 
 | | Crystal structure of the hepatitis C virus NS5B RNA-dependent RNA polymerase complex with (2E)-3-(4-{[(1-{[(13-cyclohexyl-6-oxo-6,7-dihydro-5h-indolo[1,2-d][1,4]benzodiazepin-10-yl)carbonyl]amino}cyclopentyl)carbonyl]amino}phenyl)prop-2-enoic acid | | Descriptor: | (2E)-3-(4-{[(1-{[(13-cyclohexyl-6-oxo-6,7-dihydro-5H-indolo[1,2-d][1,4]benzodiazepin-10-yl)carbonyl]amino}cyclopentyl)carbonyl]amino}phenyl)prop-2-enoic acid, RNA-directed RNA polymerase, SULFATE ION | | Authors: | Sheriff, S. | | Deposit date: | 2010-12-16 | | Release date: | 2011-04-27 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | Syntheses and initial evaluation of a series of indolo-fused heterocyclic inhibitors of the polymerase enzyme (NS5B) of the hepatitis C virus.
Bioorg.Med.Chem.Lett., 21, 2011
|
|
6IRD
 
 | | Complex structure of INADL PDZ89 and PLCb4 C-terminal CC-PBM | | Descriptor: | 1-phosphatidylinositol 4,5-bisphosphate phosphodiesterase, GOLD ION, InaD-like protein | | Authors: | Ye, F, Li, J, Huang, Y, Liu, W, Zhang, M. | | Deposit date: | 2018-11-12 | | Release date: | 2019-01-23 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.813 Å) | | Cite: | An unexpected INAD PDZ tandem-mediated plc beta binding in Drosophila photo receptors.
Elife, 7, 2018
|
|
6IRE
 
 | | Complex structure of INAD PDZ45 and NORPA CC-PBM | | Descriptor: | 1-phosphatidylinositol 4,5-bisphosphate phosphodiesterase, Inactivation-no-after-potential D protein | | Authors: | Ye, F, Li, J, Deng, X, Liu, W, Zhang, M. | | Deposit date: | 2018-11-12 | | Release date: | 2019-01-23 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (3.25 Å) | | Cite: | An unexpected INAD PDZ tandem-mediated plc beta binding in Drosophila photo receptors.
Elife, 7, 2018
|
|
6IRC
 
 | | C-terminal domain of Drosophila phospholipase b NORPA, methylated | | Descriptor: | 1-phosphatidylinositol 4,5-bisphosphate phosphodiesterase | | Authors: | Ye, F, Li, J, Huang, Y, Liu, W, Zhang, M. | | Deposit date: | 2018-11-12 | | Release date: | 2019-01-02 | | Last modified: | 2020-10-28 | | Method: | X-RAY DIFFRACTION (3.538 Å) | | Cite: | An unexpected INAD PDZ tandem-mediated plc beta binding in Drosophila photo receptors.
Elife, 7, 2018
|
|
6IRB
 
 | | C-terminal coiled coil domain of Drosophila phospholipase C beta NORPA, selenomethionine | | Descriptor: | 1-phosphatidylinositol 4,5-bisphosphate phosphodiesterase | | Authors: | Ye, F, Li, J, Huang, Y, Liu, W, Zhang, M. | | Deposit date: | 2018-11-12 | | Release date: | 2019-01-02 | | Last modified: | 2020-10-28 | | Method: | X-RAY DIFFRACTION (2.661 Å) | | Cite: | An unexpected INAD PDZ tandem-mediated plc beta binding in Drosophila photo receptors.
Elife, 7, 2018
|
|
7CQP
 
 | | Complex of TRPC4 and Calmodulin_Nlobe | | Descriptor: | CALCIUM ION, Calmodulin-1, Peptide from Short transient receptor potential channel 4 | | Authors: | Shen, Z.S. | | Deposit date: | 2020-08-11 | | Release date: | 2021-06-23 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Calmodulin binds to Drosophila TRP with an unexpected mode.
Structure, 29, 2021
|
|
7CQV
 
 | | Complex of TRP_CBS1 and Calmodulin_Nlobe | | Descriptor: | AT15141p, CALCIUM ION, Transient receptor potential protein | | Authors: | Shen, Z.S. | | Deposit date: | 2020-08-11 | | Release date: | 2021-06-23 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Calmodulin binds to Drosophila TRP with an unexpected mode.
Structure, 29, 2021
|
|
7CQH
 
 | | Complex of TRP_CBS2 and Calmodulin_Clobe | | Descriptor: | AT15141p, CALCIUM ION, Transient receptor potential protein | | Authors: | Shen, Z.S. | | Deposit date: | 2020-08-10 | | Release date: | 2021-06-23 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Calmodulin binds to Drosophila TRP with an unexpected mode.
Structure, 29, 2021
|
|
3QAU
 
 | | 3-Hydroxy-3-MethylGlutaryl-Coenzyme A Reductase from Streptococcus pneumoniae | | Descriptor: | 3-hydroxy-3-methylglutaryl-coenzyme a reductase, GLYCEROL, SODIUM ION, ... | | Authors: | Zhang, L, Feng, L, Zhou, L, Gui, J, Wan, J. | | Deposit date: | 2011-01-11 | | Release date: | 2011-01-26 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | 3-Hydroxy-3-MethylGlutaryl-Coenzyme A Reductase from Streptococcus pneumoniae
To be Published
|
|
3QAE
 
 | | 3-hydroxy-3-methylglutaryl-coenzyme A reductase of Streptococcus pneumoniae | | Descriptor: | 3-hydroxy-3-methylglutaryl-coenzyme a reductase, CITRIC ACID, GLYCEROL, ... | | Authors: | Zhang, L, Feng, L, Zhou, L, Gui, J, Wan, J. | | Deposit date: | 2011-01-10 | | Release date: | 2011-01-26 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | 3-hydroxy-3-methylglutaryl-coenzyme A reductase of Streptococcus pneumoniae
To be Published
|
|
