5TIP
 
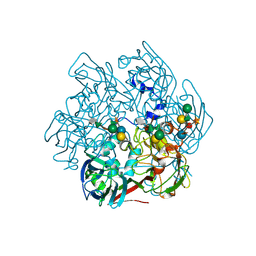 | | The Structure of the Major Capsid protein of PBCV-1 | | Descriptor: | 6-deoxy-2,3-di-O-methyl-alpha-L-mannopyranose-(1-2)-beta-L-rhamnopyranose-(1-4)-beta-D-xylopyranose-(1-4)-[alpha-D-mannopyranose-(1-3)-alpha-D-rhamnopyranose-(1-3)][alpha-D-galactopyranose-(1-2)]alpha-L-fucopyranose-(1-3)-[beta-D-xylopyranose-(1-4)]beta-D-glucopyranose, MERCURY (II) ION, Major capsid protein, ... | | Authors: | Klose, T, De Castro, C, Speciale, I, Molinaro, A, Van Etten, J.L, Rossmann, M.G. | | Deposit date: | 2016-10-03 | | Release date: | 2017-10-18 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of the chlorovirus PBCV-1 major capsid glycoprotein determined by combining crystallographic and carbohydrate molecular modeling approaches.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
5TIQ
 
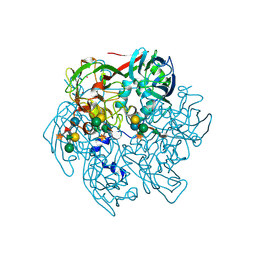 | | The Structure of the Major Capsid protein of PBCV-1 | | Descriptor: | 6-deoxy-2,3-di-O-methyl-alpha-L-mannopyranose-(1-2)-beta-L-rhamnopyranose-(1-4)-beta-D-xylopyranose-(1-4)-[alpha-D-mannopyranose-(1-3)-alpha-D-rhamnopyranose-(1-3)][alpha-D-galactopyranose-(1-2)]alpha-L-fucopyranose-(1-3)-[beta-D-xylopyranose-(1-4)]beta-D-glucopyranose, MERCURY (II) ION, Major capsid protein, ... | | Authors: | Klose, T, De Castro, C, Speciale, I, Molinaro, A, Van Etten, J.L, Rossmann, M.G. | | Deposit date: | 2016-10-03 | | Release date: | 2017-10-18 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.537 Å) | | Cite: | Structure of the chlorovirus PBCV-1 major capsid glycoprotein determined by combining crystallographic and carbohydrate molecular modeling approaches.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
6KX0
 
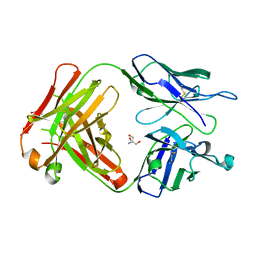 | | Crystal structure of SN-101 mAb non-liganded form | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Fab Fragment-SN-101-Heavy chain, Fab Fragment-SN-101-Light chain | | Authors: | Wakui, H, Tanaka, Y, Kato, K, Ose, T, Matsumoto, I, Min, Y, Tachibana, T, Nishimura, S.-I. | | Deposit date: | 2019-09-09 | | Release date: | 2020-07-29 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.404 Å) | | Cite: | A straightforward approach to antibodies recognising cancer specific glycopeptidic neoepitopes
Chem Sci, 11, 2020
|
|
6KX1
 
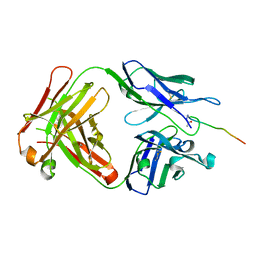 | | Crystal structure of SN-101 mAb in complex with MUC1 glycopeptide | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-galactopyranose, Fab Fragment-SN-101-Heavy chain, Fab Fragment-SN-101-Light chain, ... | | Authors: | Wakui, H, Tanaka, Y, Kato, K, Ose, T, Matsumoto, I, Min, Y, Tachibana, T, Nishimura, S.-I. | | Deposit date: | 2019-09-09 | | Release date: | 2020-07-29 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.773 Å) | | Cite: | A straightforward approach to antibodies recognising cancer specific glycopeptidic neoepitopes
Chem Sci, 11, 2020
|
|
4WCO
 
 | | Crystal structure of extracellular domain of human lectin-like transcript 1 (LLT1), the ligand for natural killer receptor-P1A | | Descriptor: | ACETATE ION, C-type lectin domain family 2 member D, SULFATE ION, ... | | Authors: | Kita, S, Matsubara, H, Kasai, Y, Tamaoki, T, Okabe, Y, Fukuhara, H, Kamishikiryo, J, Ose, T, Kuroki, K, Maenaka, K. | | Deposit date: | 2014-09-05 | | Release date: | 2015-06-24 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.46 Å) | | Cite: | Crystal structure of extracellular domain of human lectin-like transcript 1 (LLT1), the ligand for natural killer receptor-P1A
Eur.J.Immunol., 45, 2015
|
|
7XGI
 
 | | COMT SAH Mg opicapone complex | | Descriptor: | Catechol O-methyltransferase, MAGNESIUM ION, Opicapone, ... | | Authors: | Takebe, K, Kuwada-Kusunose, T, Suzuki, M, Iijima, H. | | Deposit date: | 2022-04-04 | | Release date: | 2023-04-12 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural and Computational Analyses of the Unique Interactions of Opicapone in the Binding Pocket of Catechol O -Methyltransferase: A Crystallographic Study and Fragment Molecular Orbital Analyses.
J.Chem.Inf.Model., 63, 2023
|
|
7XJB
 
 | | Rat-COMT, opicapone,SAM and Mg bond | | Descriptor: | CHLORIDE ION, Catechol O-methyltransferase, MAGNESIUM ION, ... | | Authors: | Takebe, K, Iijima, H, Suzuki, M, Kuwada-Kusunose, T. | | Deposit date: | 2022-04-15 | | Release date: | 2023-05-31 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural and Computational Analyses of the Unique Interactions of Opicapone in the Binding Pocket of Catechol O -Methyltransferase: A Crystallographic Study and Fragment Molecular Orbital Analyses.
J.Chem.Inf.Model., 63, 2023
|
|
7C21
 
 | | Crystal structure of Duvenhage virus phosphoprotein C-terminal domain | | Descriptor: | Phosphoprotein | | Authors: | Sugiyama, A, Jiang, X, Maenaka, K, Yao, M, Ose, T. | | Deposit date: | 2020-05-06 | | Release date: | 2021-03-17 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural comparison of the C-terminal domain of functionally divergent lyssavirus P proteins.
Biochem.Biophys.Res.Commun., 529, 2020
|
|
7C20
 
 | |
5XOF
 
 | | Crystal structure of human paired immunoglobulin-like type 2 receptor alpha with synthesized glycopeptide I | | Descriptor: | N-acetyl-alpha-neuraminic acid-(2-6)-2-acetamido-2-deoxy-alpha-D-glucopyranose, Paired immunoglobulin-like type 2 receptor alpha, Peptide from Nitric oxide synthase, ... | | Authors: | Furukawa, A, Kakita, K, Yamada, T, Ishizuka, M, Sakamoto, J, Hatori, N, Maeda, N, Ohsaka, F, Saitoh, T, Nomura, T, Kuroki, K, Nambu, H, Arase, H, Matsunaga, H, Anada, M, Ose, T, Hashimoto, S, Maenaka, K. | | Deposit date: | 2017-05-28 | | Release date: | 2017-10-25 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.963 Å) | | Cite: | Structural and thermodynamic analyses reveal critical features of glycopeptide recognition by the human PILR alpha immune cell receptor.
J. Biol. Chem., 292, 2017
|
|
8YBF
 
 | | Crystal structure of canine distemper virus hemagglutinin | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Hemagglutinin glycoprotein | | Authors: | Fukuhara, H, Yumoto, K, Sako, M, Kajikawa, M, Ose, T, Hashiguchi, T, Kamishikiryo, J, Maita, N, Kuroki, K, Maenaka, K. | | Deposit date: | 2024-02-13 | | Release date: | 2024-07-31 | | Last modified: | 2024-08-14 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Glycan-shielded homodimer structure and dynamical features of the canine distemper virus hemagglutinin relevant for viral entry and efficient vaccination.
Elife, 12, 2024
|
|
5XO2
 
 | | Crystal structure of human paired immunoglobulin-like type 2 receptor alpha with synthesized glycopeptide II | | Descriptor: | N-acetyl-alpha-neuraminic acid-(2-6)-2-acetamido-2,4-dideoxy-alpha-D-xylo-hexopyranose, Paired immunoglobulin-like type 2 receptor alpha, Peptide from Envelope glycoprotein B | | Authors: | Furukawa, A, Kakita, K, Yamada, T, Ishizuka, M, Sakamoto, J, Hatori, N, Maeda, N, Ohsaka, F, Saitoh, T, Nomura, T, Kuroki, K, Nambu, H, Arase, H, Matsunaga, S, Anada, M, Ose, T, Hashimoto, S, Maenaka, K. | | Deposit date: | 2017-05-25 | | Release date: | 2017-10-25 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.201 Å) | | Cite: | Structural and thermodynamic analyses reveal critical features of glycopeptide recognition by the human PILR alpha immune cell receptor.
J. Biol. Chem., 292, 2017
|
|
4F86
 
 | | Structure analysis of Geranyl diphosphate methyltransferase in complex with GPP and sinefungin | | Descriptor: | GERANYL DIPHOSPHATE, Geranyl diphosphate 2-C-methyltransferase, MAGNESIUM ION, ... | | Authors: | Ariyawutthiphan, O, Ose, T, Minami, A, Gao, Y.G, Yao, M, Oikawa, H, Tanaka, I. | | Deposit date: | 2012-05-17 | | Release date: | 2012-10-31 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structure analysis of geranyl pyrophosphate methyltransferase and the proposed reaction mechanism of SAM-dependent C-methylation
Acta Crystallogr.,Sect.D, 68, 2012
|
|
4F85
 
 | | Structure analysis of Geranyl diphosphate methyltransferase | | Descriptor: | Geranyl diphosphate 2-C-methyltransferase | | Authors: | Ariyawutthiphan, O, Ose, T, Minami, A, Gao, Y.G, Yao, M, Oikawa, H, Tanaka, I. | | Deposit date: | 2012-05-17 | | Release date: | 2012-10-31 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure analysis of geranyl pyrophosphate methyltransferase and the proposed reaction mechanism of SAM-dependent C-methylation
Acta Crystallogr.,Sect.D, 68, 2012
|
|
4F84
 
 | | Structure analysis of Geranyl diphosphate methyltransferase in complex with SAM | | Descriptor: | Geranyl diphosphate 2-C-methyltransferase, S-ADENOSYLMETHIONINE | | Authors: | Ariyawutthiphan, O, Ose, T, Minami, A, Gao, Y.G, Yao, M, Oikawa, H, Tanaka, I. | | Deposit date: | 2012-05-17 | | Release date: | 2012-10-31 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure analysis of geranyl pyrophosphate methyltransferase and the proposed reaction mechanism of SAM-dependent C-methylation
Acta Crystallogr.,Sect.D, 68, 2012
|
|
6A97
 
 | | Crystal structure of MHC-like MILL2 | | Descriptor: | Beta-2-microglobulin, MHC I-like leukocyte 2 long form, SULFATE ION | | Authors: | Kajikawa, M, Ose, T, Maenaka, K. | | Deposit date: | 2018-07-11 | | Release date: | 2018-12-05 | | Method: | X-RAY DIFFRACTION (2.148 Å) | | Cite: | Structure of MHC class I-like MILL2 reveals heparan-sulfate binding and interdomain flexibility.
Nat Commun, 9, 2018
|
|
8YIE
 
 | | Crystal structure of GH13_30 alpha-glucosidase CmmB in complex with acarbose | | Descriptor: | 4,6-dideoxy-4-{[(1S,4R,5S,6S)-4,5,6-trihydroxy-3-(hydroxymethyl)cyclohex-2-en-1-yl]amino}-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose, Alpha-glucosidase | | Authors: | Saburi, W, Tagami, T, Yu, J, Ose, T, Yao, M, Mori, H. | | Deposit date: | 2024-02-29 | | Release date: | 2024-07-03 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Molecular mechanism for the substrate specificity of Arthrobacter globiformis M6 alpha-glucosidase CmmB, belonging to glycoside hydrolase family 13 subfamily 30
Food Biosci, 61, 2024
|
|
8YIF
 
 | | Crystal structure of GH13_30 alpha-glucosidase CmmB in complex with acarviosin | | Descriptor: | Acarviosin, Alpha-glucosidase | | Authors: | Saburi, W, Tagami, T, Yu, J, Ose, T, Yao, M, Mori, H. | | Deposit date: | 2024-02-29 | | Release date: | 2024-07-03 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Molecular mechanism for the substrate specificity of Arthrobacter globiformis M6 alpha-glucosidase CmmB, belonging to glycoside hydrolase family 13 subfamily 30
Food Biosci, 61, 2024
|
|
1WOC
 
 | | Crystal structure of PriB | | Descriptor: | Primosomal replication protein n | | Authors: | Shioi, S, Ose, T, Maenaka, K, Abe, Y, Kohda, D, Katayama, T, Ueda, T. | | Deposit date: | 2004-08-13 | | Release date: | 2005-01-25 | | Last modified: | 2012-12-05 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of a biologically functional form of PriB from Escherichia coli reveals a potential single-stranded DNA-binding site
Biochem.Biophys.Res.Commun., 326, 2005
|
|
8HRH
 
 | | SN-131/1B2 anti-MUC1 antibody with a glycopeptide | | Descriptor: | 1-ACETYL-L-PROLINE, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ALANINE, ... | | Authors: | Wakui, H, Horidome, C, Yao, M, Ose, T, Nishimura, S.-I. | | Deposit date: | 2022-12-15 | | Release date: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Structural and molecular insight into antibody recognition of dynamic neoepitopes in membrane tethered MUC1 of pancreatic cancer cells and secreted exosomes.
Rsc Chem Biol, 4, 2023
|
|
1WSU
 
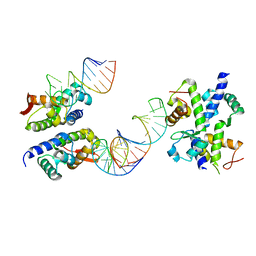 | | C-terminal domain of elongation factor selB complexed with SECIS RNA | | Descriptor: | 5'-R(*GP*GP*CP*GP*UP*UP*GP*CP*CP*GP*GP*UP*CP*U*GP*GP*CP*AP*AP*CP*GP*CP*C)-3', Selenocysteine-specific elongation factor | | Authors: | Yoshizawa, S, Rasubala, L, Ose, T, Kohda, D, Fourmy, D, Maenaka, K. | | Deposit date: | 2004-11-11 | | Release date: | 2005-01-25 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural basis for mRNA recognition by elongation factor SelB
Nat.Struct.Mol.Biol., 12, 2005
|
|
6IMF
 
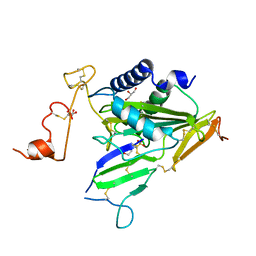 | | Crystal structure of TOXIN/ANTITOXIN complex | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Cysteine-rich venom protein triflin, GLYCEROL, ... | | Authors: | Shioi, N, Tadokoro, T, Shioi, S, Hu, Y, Kurahara, L.H, Okabe, Y, Matsubara, H, Kita, S, Ose, T, Kuroki, K, Maenaka, K, Terada, S. | | Deposit date: | 2018-10-22 | | Release date: | 2018-12-12 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of the complex between venom toxin and serum inhibitor from Viperidae snake.
J. Biol. Chem., 294, 2019
|
|
6K6N
 
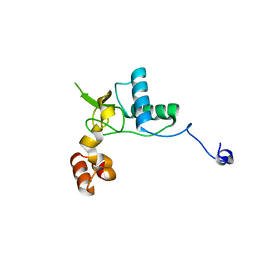 | | Crystal structure of SIVmac239 Nef protein | | Descriptor: | Protein Nef | | Authors: | Hirao, K, Andrews, S, Kuroki, K, Kusaka, H, Tadokoro, T, Kita, S, Ose, T, Rowland-Jones, S, Maenaka, K. | | Deposit date: | 2019-06-04 | | Release date: | 2020-03-25 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.0002 Å) | | Cite: | Structure of HIV-2 Nef Reveals Features Distinct from HIV-1 Involved in Immune Regulation.
Iscience, 23, 2020
|
|
6K60
 
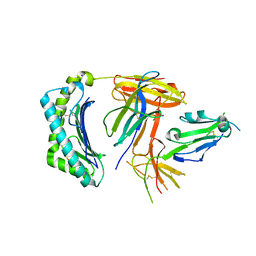 | | Structural and functional basis for HLA-G isoform recognition of immune checkpoint receptor LILRBs | | Descriptor: | Beta-2-microglobulin, HLA class I histocompatibility antigen, alpha chain G, ... | | Authors: | Kuroki, K, Matsubara, H, Kanda, R, Miyashita, N, Shiroishi, M, Fukunaga, Y, Kamishikiryo, J, Fukunaga, A, Hirose, K, Sugita, Y, Kita, S, Ose, T, Maenaka, K. | | Deposit date: | 2019-05-31 | | Release date: | 2019-11-27 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.149 Å) | | Cite: | Structural and Functional Basis for LILRB Immune Checkpoint Receptor Recognition of HLA-G Isoforms.
J Immunol., 203, 2019
|
|
6K6M
 
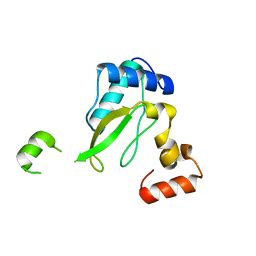 | | Crystal structure of HIV-2 Nef protein | | Descriptor: | Protein Nef | | Authors: | Hirao, K, Andrews, S, Kuroki, K, Kusaka, H, Tadokoro, T, Kita, S, Ose, T, Rowland-Jones, S, Maenaka, K. | | Deposit date: | 2019-06-04 | | Release date: | 2020-03-25 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.072 Å) | | Cite: | Structure of HIV-2 Nef Reveals Features Distinct from HIV-1 Involved in Immune Regulation.
Iscience, 23, 2020
|
|
