2AME
 
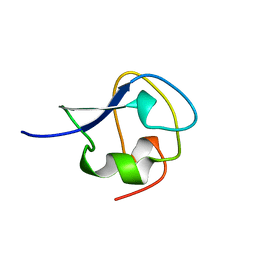 | | TYPE III ANTIFREEZE PROTEIN ISOFORM HPLC 12 N14Q | | Descriptor: | PROTEIN (ANTIFREEZE PROTEIN TYPE III) | | Authors: | Graether, S.P, Deluca, C.I, Baardsnes, J, Hill, G.A, Davies, P.L, Jia, Z. | | Deposit date: | 1999-01-18 | | Release date: | 1999-04-30 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Quantitative and qualitative analysis of type III antifreeze protein structure and function.
J.Biol.Chem., 274, 1999
|
|
7AME
 
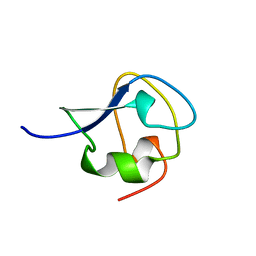 | | TYPE III ANTIFREEZE PROTEIN ISOFORM HPLC 12 T15A | | Descriptor: | PROTEIN (ANTIFREEZE PROTEIN TYPE III) | | Authors: | Graether, S.P, Deluca, C.I, Baardsnes, J, Hill, G.A, Davies, P.L, Jia, Z. | | Deposit date: | 1999-01-24 | | Release date: | 1999-04-29 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Quantitative and qualitative analysis of type III antifreeze protein structure and function.
J.Biol.Chem., 274, 1999
|
|
5VTM
 
 | |
9MSI
 
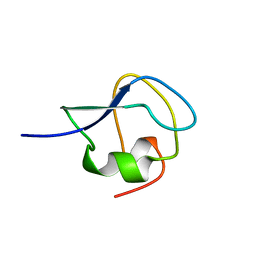 | | TYPE III ANTIFREEZE PROTEIN ISOFORM HPLC 12 T18N | | Descriptor: | PROTEIN (ANTIFREEZE PROTEIN TYPE III) | | Authors: | Graether, S.P, Deluca, C.I, Baardsnes, J, Hill, G.A, Davies, P.L, Jia, Z. | | Deposit date: | 1999-01-24 | | Release date: | 1999-04-28 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Quantitative and qualitative analysis of type III antifreeze protein structure and function.
J.Biol.Chem., 274, 1999
|
|
2SPG
 
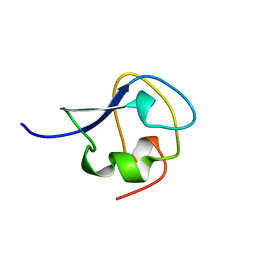 | | TYPE III ANTIFREEZE PROTEIN ISOFORM HPLC 12 T15S | | Descriptor: | PROTEIN (ANTIFREEZE PROTEIN TYPE III) | | Authors: | Graether, S.P, Deluca, C.I, Baardsnes, J, Hill, G.A, Davies, P.L, Jia, Z. | | Deposit date: | 1999-01-21 | | Release date: | 1999-04-28 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Quantitative and qualitative analysis of type III antifreeze protein structure and function.
J.Biol.Chem., 274, 1999
|
|
9AME
 
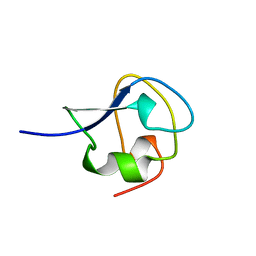 | | TYPE III ANTIFREEZE PROTEIN ISOFORM HPLC 12 S42G | | Descriptor: | PROTEIN (ANTIFREEZE PROTEIN TYPE III) | | Authors: | Graether, S.P, Deluca, C.I, Baardsnes, J, Hill, G.A, Davies, P.L, Jia, Z. | | Deposit date: | 1999-01-24 | | Release date: | 1999-04-29 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Quantitative and qualitative analysis of type III antifreeze protein structure and function.
J.Biol.Chem., 274, 1999
|
|
1MSJ
 
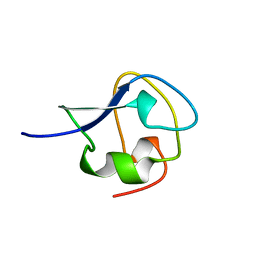 | | TYPE III ANTIFREEZE PROTEIN ISOFORM HPLC 12 T15V | | Descriptor: | PROTEIN (ANTIFREEZE PROTEIN TYPE III) | | Authors: | Graether, S.P, Deluca, C.I, Baardsnes, J, Hill, G.A, Davies, P.L, Jia, Z. | | Deposit date: | 1999-01-24 | | Release date: | 1999-04-29 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Quantitative and qualitative analysis of type III antifreeze protein structure and function.
J.Biol.Chem., 274, 1999
|
|
1JAB
 
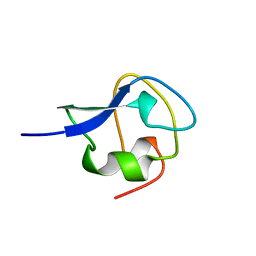 | | TYPE III ANTIFREEZE PROTEIN ISOFORM HPLC 12 T18S | | Descriptor: | PROTEIN (ANTIFREEZE PROTEIN TYPE III) | | Authors: | Graether, S.P, Deluca, C.I, Baardsnes, J, Hill, G.A, Davies, P.L, Jia, Z. | | Deposit date: | 1999-01-24 | | Release date: | 1999-04-29 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Quantitative and qualitative analysis of type III antifreeze protein structure and function.
J.Biol.Chem., 274, 1999
|
|
3AME
 
 | | TYPE III ANTIFREEZE PROTEIN ISOFORM HPLC 12 Q9TQ44T | | Descriptor: | PROTEIN (ANTIFREEZE PROTEIN TYPE III) | | Authors: | Graether, S.P, Deluca, C.I, Baardsnes, J, Hill, G.A, Davies, P.L, Jia, Z. | | Deposit date: | 1999-01-24 | | Release date: | 1999-04-29 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Quantitative and qualitative analysis of type III antifreeze protein structure and function.
J.Biol.Chem., 274, 1999
|
|
1B7J
 
 | | TYPE III ANTIFREEZE PROTEIN ISOFORM HPLC 12 V20A | | Descriptor: | PROTEIN (ANTIFREEZE PROTEIN TYPE III) | | Authors: | Graether, S.P, Deluca, C.I, Baardsnes, J, Hill, G.A, Davies, P.L, Jia, Z. | | Deposit date: | 1999-01-24 | | Release date: | 1999-04-30 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Quantitative and qualitative analysis of type III antifreeze protein structure and function.
J.Biol.Chem., 274, 1999
|
|
5MSI
 
 | |
1EKL
 
 | | TYPE III ANTIFREEZE PROTEIN ISOFORM HPLC 12 E35K | | Descriptor: | PROTEIN (ANTIFREEZE PROTEIN TYPE III) | | Authors: | Graether, S.P, Deluca, C.I, Baardsnes, J, Hill, G.A, Davies, P.L, Jia, Z. | | Deposit date: | 1999-01-21 | | Release date: | 1999-04-29 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Quantitative and qualitative analysis of type III antifreeze protein structure and function.
J.Biol.Chem., 274, 1999
|
|
7MSI
 
 | |
1MDW
 
 | | Crystal Structure of Calcium-Bound Protease Core of Calpain II Reveals the Basis for Intrinsic Inactivation | | Descriptor: | CALCIUM ION, Calpain II, catalytic subunit | | Authors: | Moldoveanu, T, Hosfield, C.M, Lim, D, Jia, Z, Davies, P.L. | | Deposit date: | 2002-08-07 | | Release date: | 2003-04-29 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Calpain silencing by a reversible intrinsic mechanism.
Nat.Struct.Biol., 10, 2003
|
|
4CDP
 
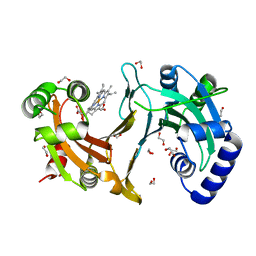 | | Improved coordinates for Escherichia coli O157:H7 heme degrading enzyme ChuS. | | Descriptor: | 1,2-ETHANEDIOL, FORMIC ACID, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Suits, M.D.L, Jaffer, N, Jia, Z. | | Deposit date: | 2013-11-05 | | Release date: | 2013-11-13 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Structure of the Escherichia Coli O157:H7 Heme Oxygenase Chus in Complex with Heme and Enzymatic Inactivation by Mutation of the Heme Coordinating Residue His-193.
J.Biol.Chem., 281, 2006
|
|
1N4I
 
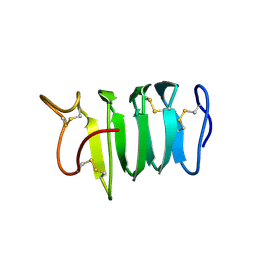 | | Solution structure of spruce budworm antifreeze protein at 5 degrees celsius | | Descriptor: | thermal hysteresis protein | | Authors: | Graether, S.P, Gagne, S.M, Spyracopoulos, L, Jia, Z, Davies, P.L, Sykes, B.D. | | Deposit date: | 2002-10-31 | | Release date: | 2003-04-08 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | Spruce Budworm Antifreeze Protein: Changes in Structure and Dynamics at Low Temperature
J.Mol.Biol., 327, 2003
|
|
1ZYL
 
 | |
2B3D
 
 | |
2AMJ
 
 | |
1DKM
 
 | | CRYSTAL STRUCTURE OF ESCHERICHIA COLI PHYTASE AT PH 6.6 WITH HG2+ CATION ACTING AS AN INTERMOLECULAR BRIDGE | | Descriptor: | MERCURY (II) ION, PHYTASE | | Authors: | Lim, D, Golovan, S, Forsberg, C.W, Jia, Z. | | Deposit date: | 1999-12-08 | | Release date: | 2000-08-02 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Crystal structures of Escherichia coli phytase and its complex with phytate.
Nat.Struct.Biol., 7, 2000
|
|
1DKQ
 
 | | CRYSTAL STRUCTURE OF PHYTATE COMPLEX ESCHERICHIA COLI PHYTASE AT PH 5.0. PHYTATE IS BOUND WITH ITS 3-PHOSPHATE IN THE ACTIVE SITE. HG2+ CATION ACTS AS AN INTERMOLECULAR BRIDGE | | Descriptor: | INOSITOL HEXAKISPHOSPHATE, MERCURY (II) ION, PHYTASE | | Authors: | Lim, D, Golovan, S, Forsberg, C.W, Jia, Z. | | Deposit date: | 1999-12-08 | | Release date: | 2000-08-03 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Crystal structures of Escherichia coli phytase and its complex with phytate.
Nat.Struct.Biol., 7, 2000
|
|
1DKN
 
 | | CRYSTAL STRUCTURE OF ESCHERICHIA COLI PHYTASE AT PH 5.0 WITH HG2+ CATION ACTING AS AN INTERMOLECULAR BRIDGE | | Descriptor: | MERCURY (II) ION, PHYTASE | | Authors: | Lim, D, Golovan, S, Forsberg, C.W, Jia, Z. | | Deposit date: | 1999-12-08 | | Release date: | 2000-08-03 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structures of Escherichia coli phytase and its complex with phytate.
Nat.Struct.Biol., 7, 2000
|
|
1DKP
 
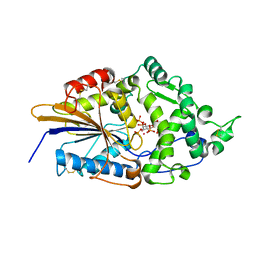 | | CRYSTAL STRUCTURE OF PHYTATE COMPLEX OF ESCHERICHIA COLI PHYTASE AT PH 6.6. PHYTATE IS BOUND WITH ITS 3-PHOSPHATE IN THE ACTIVE SITE. HG2+ CATION ACTS AS AN INTERMOLECULAR BRIDGE | | Descriptor: | INOSITOL HEXAKISPHOSPHATE, MERCURY (II) ION, PHYTASE | | Authors: | Lim, D, Golovan, S, Forsberg, C.W, Jia, Z. | | Deposit date: | 1999-12-08 | | Release date: | 2000-08-03 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Crystal structures of Escherichia coli phytase and its complex with phytate.
Nat.Struct.Biol., 7, 2000
|
|
1DF0
 
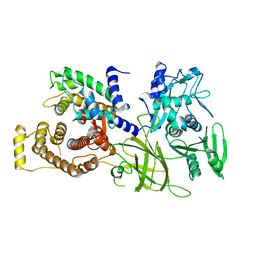 | | Crystal structure of M-Calpain | | Descriptor: | CALPAIN, M-CALPAIN | | Authors: | Hosfield, C.M, Elce, J.S, Davies, P.L, Jia, Z. | | Deposit date: | 1999-11-16 | | Release date: | 2000-06-21 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of calpain reveals the structural basis for Ca(2+)-dependent protease activity and a novel mode of enzyme activation.
EMBO J., 18, 1999
|
|
1DKL
 
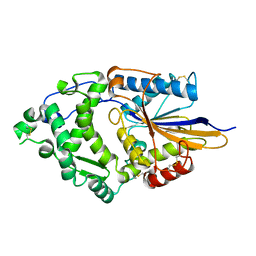 | |
