8PR7
 
 | |
8BMV
 
 | | Ligand binding domain of the P. Putida receptor McpH in complex with Uric acid | | Descriptor: | Methyl-accepting chemotaxis protein McpH, URIC ACID | | Authors: | Gavira, J.A, Krell, T, Fernandez, M, Martinez-Rodriguez, S. | | Deposit date: | 2022-11-11 | | Release date: | 2024-07-24 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Ubiquitous purine sensor modulates diverse signal transduction pathways in bacteria.
Nat Commun, 15, 2024
|
|
1A2S
 
 | | THE SOLUTION NMR STRUCTURE OF OXIDIZED CYTOCHROME C6 FROM THE GREEN ALGA MONORAPHIDIUM BRAUNII, MINIMIZED AVERAGE STRUCTURE | | Descriptor: | CYTOCHROME C6, HEME C | | Authors: | Banci, L, Bertini, I, De La Rosa, M.A, Koulougliotis, D, Navarro, J.A, Walter, O. | | Deposit date: | 1998-01-10 | | Release date: | 1998-04-29 | | Last modified: | 2022-02-16 | | Method: | SOLUTION NMR | | Cite: | Solution structure of oxidized cytochrome c6 from the green alga Monoraphidium braunii.
Biochemistry, 37, 1998
|
|
154D
 
 | | DNA DISTORTION IN BIS-INTERCALATED COMPLEXES | | Descriptor: | BIS-(N-ETHYLPYRIDINIUM-(3-METHOXYCARBAZOLE))HEXANE-1,6-DIAMINE, DNA (5'-D(*(CBR)P*GP*CP*G)-3') | | Authors: | Peek, M.E, Lipscomb, L.A, Bertrand, J.A, Gao, Q, Roques, B.P, Garbay-Jaureguiberry, C, Williams, L.D. | | Deposit date: | 1993-12-14 | | Release date: | 1994-08-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | DNA distortion in bis-intercalated complexes.
Biochemistry, 33, 1994
|
|
1T39
 
 | | HUMAN O6-ALKYLGUANINE-DNA ALKYLTRANSFERASE COVALENTLY CROSSLINKED TO DNA | | Descriptor: | 5'-D(*GP*CP*CP*AP*TP*GP*(E1X)P*CP*TP*AP*GP*TP*A)-3', 5'-D(*TP*AP*CP*TP*AP*GP*CP*CP*AP*TP*GP*GP*C)-3', Methylated-DNA--protein-cysteine methyltransferase | | Authors: | Daniels, D.S, Woo, T.T, Luu, K.X, Noll, D.M, Clarke, N.D, Pegg, A.E, Tainer, J.A. | | Deposit date: | 2004-04-25 | | Release date: | 2004-07-13 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | DNA binding and nucleotide flipping by the human DNA repair protein AGT.
Nat.Struct.Mol.Biol., 11, 2004
|
|
1T38
 
 | | HUMAN O6-ALKYLGUANINE-DNA ALKYLTRANSFERASE BOUND TO DNA CONTAINING O6-METHYLGUANINE | | Descriptor: | 5'-D(*GP*CP*CP*AP*TP*GP*(6OG)P*CP*TP*AP*GP*TP*A)-3', 5'-D(*TP*AP*CP*TP*AP*GP*CP*CP*AP*TP*GP*GP*C)-3', Methylated-DNA--protein-cysteine methyltransferase | | Authors: | Daniels, D.S, Woo, T.T, Luu, K.X, Noll, D.M, Clarke, N.D, Pegg, A.E, Tainer, J.A. | | Deposit date: | 2004-04-25 | | Release date: | 2004-07-13 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | DNA binding and nucleotide flipping by the human DNA repair protein AGT.
Nat.Struct.Mol.Biol., 11, 2004
|
|
2HPD
 
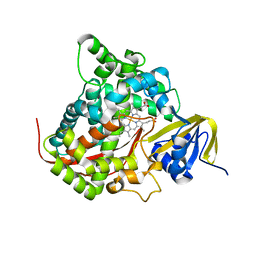 | | CRYSTAL STRUCTURE OF HEMOPROTEIN DOMAIN OF P450BM-3, A PROTOTYPE FOR MICROSOMAL P450'S | | Descriptor: | CYTOCHROME P450 BM-3, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Ravichandran, K.G, Boddupalli, S.S, Hasemann, C.A, Peterson, J.A, Deisenhofer, J. | | Deposit date: | 1993-09-16 | | Release date: | 1993-10-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of hemoprotein domain of P450BM-3, a prototype for microsomal P450's.
Science, 261, 1993
|
|
7NZQ
 
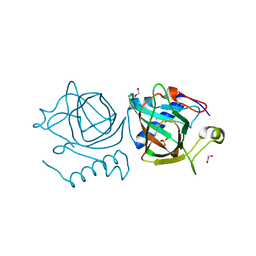 | | D-lyxose isomerase from the hyperthermophilic archaeon Thermofilum sp complexed with D-mannose | | Descriptor: | 1,2-ETHANEDIOL, D-lyxose/D-mannose family sugar isomerase, DI(HYDROXYETHYL)ETHER, ... | | Authors: | De Rose, S.A, Isupov, M.N, Littlechild, J.A, Schoenheit, P. | | Deposit date: | 2021-03-24 | | Release date: | 2021-10-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | Biochemical and Structural Characterisation of a Novel D-Lyxose Isomerase From the Hyperthermophilic Archaeon Thermofilum sp.
Front Bioeng Biotechnol, 9, 2021
|
|
7NZO
 
 | | D-lyxose isomerasefrom the hyperthermophilic archaeon Thermofilum sp | | Descriptor: | 1,2-ETHANEDIOL, D-lyxose/D-mannose family sugar isomerase, MANGANESE (II) ION | | Authors: | De Rose, S.A, Isupov, M.N, Littlechild, J.A, Schoenheit, P. | | Deposit date: | 2021-03-24 | | Release date: | 2021-10-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Biochemical and Structural Characterisation of a Novel D-Lyxose Isomerase From the Hyperthermophilic Archaeon Thermofilum sp.
Front Bioeng Biotechnol, 9, 2021
|
|
7NZP
 
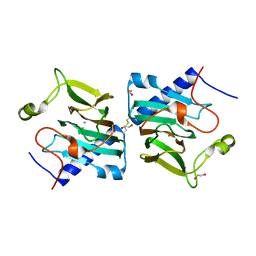 | | D-lyxose isomerase from the hyperthermophilic archaeon Thermofilum sp complexed with D-fructose | | Descriptor: | 1,2-ETHANEDIOL, D-lyxose/D-mannose family sugar isomerase, MANGANESE (II) ION, ... | | Authors: | De Rose, S.A, Isupov, M.N, Littlechild, J.A, Schoenheit, P. | | Deposit date: | 2021-03-24 | | Release date: | 2021-10-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.345 Å) | | Cite: | Biochemical and Structural Characterisation of a Novel D-Lyxose Isomerase From the Hyperthermophilic Archaeon Thermofilum sp.
Front Bioeng Biotechnol, 9, 2021
|
|
7O4A
 
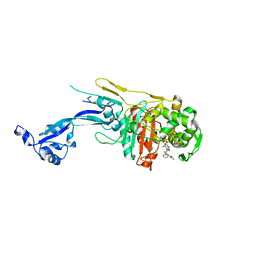 | |
7O49
 
 | |
7O4C
 
 | |
5K97
 
 | | Flap endonuclease 1 (FEN1) D233N with cleaved product fragment and Sm3+ | | Descriptor: | 1,2-ETHANEDIOL, DNA (5'-D(*AP*CP*CP*GP*TP*CP*C)-3'), DNA (5'-D(*AP*CP*TP*CP*TP*GP*CP*CP*TP*CP*AP*AP*GP*AP*CP*GP*GP*T)-3'), ... | | Authors: | Tsutakawa, S.E, Arvai, A.S, Tainer, J.A. | | Deposit date: | 2016-05-31 | | Release date: | 2017-06-28 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.102 Å) | | Cite: | Phosphate steering by Flap Endonuclease 1 promotes 5'-flap specificity and incision to prevent genome instability.
Nat Commun, 8, 2017
|
|
1JHK
 
 | |
1JPZ
 
 | | Crystal structure of a complex of the heme domain of P450BM-3 with N-Palmitoylglycine | | Descriptor: | BIFUNCTIONAL P-450:NADPH-P450 REDUCTASE, N-PALMITOYLGLYCINE, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Haines, D.C, Tomchick, D.R, Machius, M, Peterson, J.A. | | Deposit date: | 2001-08-03 | | Release date: | 2001-11-09 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Pivotal role of water in the mechanism of P450BM-3.
Biochemistry, 40, 2001
|
|
2VNJ
 
 | |
2WC1
 
 | | Three-dimensional Structure of the Nitrogen Fixation Flavodoxin (NifF) from Rhodobacter capsulatus at 2.2 A | | Descriptor: | FLAVIN MONONUCLEOTIDE, FLAVODOXIN | | Authors: | Perez-Dorado, I, Bittel, C, Hermoso, J.A, Cortez, N, Carrillo, N. | | Deposit date: | 2009-03-06 | | Release date: | 2010-04-21 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | Structural and Phylogenetic Analysis of Rhodobacter Capsulatus Niff: Uncovering General Features of Nitrogen-Fixation (Nif)-Flavodoxins.
Int.J.Mol.Sci., 14, 2013
|
|
6I4O
 
 | |
6J7O
 
 | | Crystal structure of toxin TglT (unusual type guanylyltransferase-like toxin, Rv1045) mutant E146Q from Mycobacterium tuberculosis | | Descriptor: | MAGNESIUM ION, guanylyltransferase-like toxin | | Authors: | Yu, X, Gao, X, Zhu, K, Wojdyla, J.A, Wang, M, Cui, S. | | Deposit date: | 2019-01-18 | | Release date: | 2020-05-13 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Characterization of a toxin-antitoxin system in Mycobacterium tuberculosis suggests neutralization by phosphorylation as the antitoxicity mechanism.
Commun Biol, 3, 2020
|
|
6J7T
 
 | | Crystal structure of toxin TglT (unusual type guanylyltransferase-like toxin, Rv1045) mutant D82A from Mycobacterium tuberculosis | | Descriptor: | GLYCEROL, MAGNESIUM ION, guanylyltransferase-like toxin | | Authors: | Yu, X, Gao, X, Zhu, K, Wojdyla, J.A, Wang, M, Cui, S. | | Deposit date: | 2019-01-18 | | Release date: | 2020-05-13 | | Last modified: | 2020-05-20 | | Method: | X-RAY DIFFRACTION (1.903 Å) | | Cite: | Characterization of a toxin-antitoxin system in Mycobacterium tuberculosis suggests neutralization by phosphorylation as the antitoxicity mechanism.
Commun Biol, 3, 2020
|
|
6J7R
 
 | | Crystal structure of toxin TglT (unusual type guanylyltransferase-like toxin, Rv1045) mutant S78A co-expressed with TakA from Mycobacterium tuberculosis | | Descriptor: | MAGNESIUM ION, guanylyltransferase-like toxin | | Authors: | Yu, X, Gao, X, Zhu, K, Wojdyla, J.A, Wang, M, Cui, S. | | Deposit date: | 2019-01-18 | | Release date: | 2020-05-13 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.299 Å) | | Cite: | Characterization of a toxin-antitoxin system in Mycobacterium tuberculosis suggests neutralization by phosphorylation as the antitoxicity mechanism.
Commun Biol, 3, 2020
|
|
6J7N
 
 | | Crystal structure of toxin TglT (unusual type guanylyltransferase-like toxin, Rv1045) mutant D82A co-expressed with TakA from Mycobacterium tuberculosis | | Descriptor: | MAGNESIUM ION, guanylyltransferase-like toxin | | Authors: | Yu, X, Gao, X, Zhu, K, Wojdyla, J.A, Wang, M, Cui, S. | | Deposit date: | 2019-01-18 | | Release date: | 2020-05-13 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.294 Å) | | Cite: | Characterization of a toxin-antitoxin system in Mycobacterium tuberculosis suggests neutralization by phosphorylation as the antitoxicity mechanism.
Commun Biol, 3, 2020
|
|
6J7S
 
 | | Crystal structure of toxin TglT (unusual type guanylyltransferase-like toxin, Rv1045) wild type protein from Mycobacterium tuberculosis | | Descriptor: | MAGNESIUM ION, guanylyltransferase-like toxin | | Authors: | Yu, X, Gao, X, Zhu, K, Wojdyla, J.A, Wang, M, Cui, S. | | Deposit date: | 2019-01-18 | | Release date: | 2020-05-13 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.102 Å) | | Cite: | Characterization of a toxin-antitoxin system in Mycobacterium tuberculosis suggests neutralization by phosphorylation as the antitoxicity mechanism.
Commun Biol, 3, 2020
|
|
8TUK
 
 | | Alvinella ASCC1 KH and Phosphodiesterase/Ligase Domain | | Descriptor: | 1,2-ETHANEDIOL, Activating signal cointegrator 1 complex subunit 1, IMIDAZOLE | | Authors: | Tsutakawa, S.E, Tainer, J.A, Arvai, A.S, Chinnam, N.B. | | Deposit date: | 2023-08-16 | | Release date: | 2024-07-10 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | ASCC1 structures and bioinformatics reveal a novel helix-clasp-helix RNA-binding motif linked to a two-histidine phosphodiesterase.
J.Biol.Chem., 300, 2024
|
|
