6ZHE
 
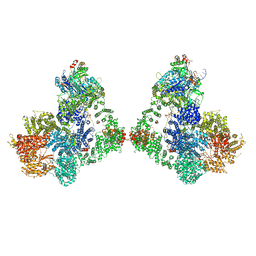 | | Cryo-EM structure of DNA-PK dimer | | Descriptor: | DNA (25-MER), DNA (26-MER), DNA (27-MER), ... | | Authors: | Chaplin, A.K, Hardwick, S.W, Chirgadze, D.Y, Blundell, T.L. | | Deposit date: | 2020-06-23 | | Release date: | 2020-10-21 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (7.24 Å) | | Cite: | Dimers of DNA-PK create a stage for DNA double-strand break repair.
Nat.Struct.Mol.Biol., 28, 2021
|
|
6ZH2
 
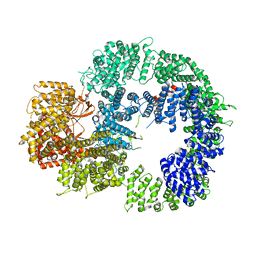 | | Cryo-EM structure of DNA-PKcs (State 1) | | Descriptor: | DNA-dependent protein kinase catalytic subunit,DNA-dependent protein kinase catalytic subunit,DNA-dependent protein kinase catalytic subunit,DNA-dependent protein kinase catalytic subunit,DNA-dependent protein kinase catalytic subunit,DNA-dependent protein kinase catalytic subunit,DNA-dependent protein kinase catalytic subunit,DNA-dependent protein kinase catalytic subunit,DNA-PKcs | | Authors: | Chaplin, A.K, Hardwick, S.W, Chirgadze, D.Y, Blundell, T.L. | | Deposit date: | 2020-06-20 | | Release date: | 2020-10-21 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (3.92 Å) | | Cite: | Dimers of DNA-PK create a stage for DNA double-strand break repair.
Nat.Struct.Mol.Biol., 28, 2021
|
|
6ZH4
 
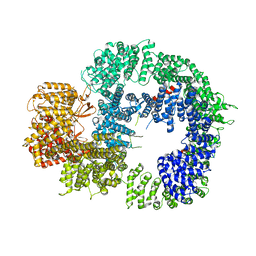 | | Cryo-EM structure of DNA-PKcs (State 3) | | Descriptor: | DNA-dependent protein kinase catalytic subunit,DNA-PKcs | | Authors: | Chaplin, A.K, Hardwick, S.W, Chirgadze, D.Y, Blundell, T.L. | | Deposit date: | 2020-06-20 | | Release date: | 2020-10-21 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (3.62 Å) | | Cite: | Dimers of DNA-PK create a stage for DNA double-strand break repair.
Nat.Struct.Mol.Biol., 28, 2021
|
|
6ZH6
 
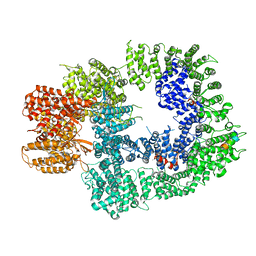 | | Cryo-EM structure of DNA-PKcs:Ku80ct194 | | Descriptor: | DNA-dependent protein kinase catalytic subunit,DNA-PKcs, X-ray repair cross-complementing protein 5 | | Authors: | Chaplin, A.K, Hardwick, S.W, Chirgadze, D.Y, Blundell, T.L. | | Deposit date: | 2020-06-21 | | Release date: | 2020-10-21 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (3.93 Å) | | Cite: | Dimers of DNA-PK create a stage for DNA double-strand break repair.
Nat.Struct.Mol.Biol., 28, 2021
|
|
4ER1
 
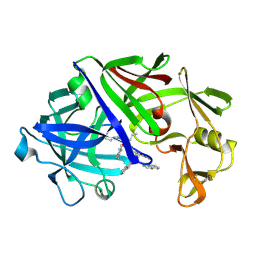 | | THE ACTIVE SITE OF ASPARTIC PROTEINASES | | Descriptor: | ENDOTHIAPEPSIN, N-[(1R,2R,4R)-1-(cyclohexylmethyl)-2-hydroxy-6-methyl-4-{[(2R)-2-methylbutyl]carbamoyl}heptyl]-3-(1H-imidazol-3-ium-4-y l)-N~2~-[3-naphthalen-1-yl-2-(naphthalen-1-ylmethyl)propanoyl]-L-alaninamide | | Authors: | Quail, J.W, Cooper, J.B, Szelke, M, Blundell, T.L. | | Deposit date: | 1990-10-14 | | Release date: | 1991-01-15 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The active site of aspartic proteinases
FEBS Lett., 174, 1984
|
|
4ER2
 
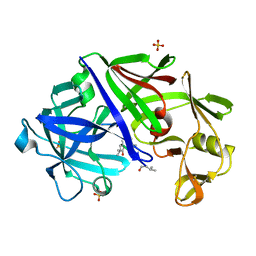 | | The active site of aspartic proteinases | | Descriptor: | ENDOTHIAPEPSIN, PEPSTATIN, SULFATE ION | | Authors: | Bailey, D, Veerapandian, B, Cooper, J.B, Blundell, T.L. | | Deposit date: | 1990-10-20 | | Release date: | 1991-01-15 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The active site of aspartic proteinases
FEBS Lett., 174, 1984
|
|
5ER2
 
 | | High-resolution X-ray diffraction study of the complex between endothiapepsin and an oligopeptide inhibitor. the analysis of the inhibitor binding and description of the rigid body shift in the enzyme | | Descriptor: | 6-ammonio-N-{[(2R,3R)-3-{[N-(tert-butoxycarbonyl)-L-phenylalanyl-3-(1H-imidazol-3-ium-4-yl)-L-alanyl]amino}-4-cyclohexyl-2-hydroxybutyl](2-methylpropyl)carbamoyl}-L-norleucyl-L-phenylalanine, ENDOTHIAPEPSIN | | Authors: | Sali, A, Veerapandian, B, Cooper, J.B, Foundling, S.I, Hoover, D.J, Blundell, T.L. | | Deposit date: | 1991-01-02 | | Release date: | 1991-04-15 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | High-resolution X-ray diffraction study of the complex between endothiapepsin and an oligopeptide inhibitor: the analysis of the inhibitor binding and description of the rigid body shift in the enzyme.
EMBO J., 8, 1989
|
|
5ER1
 
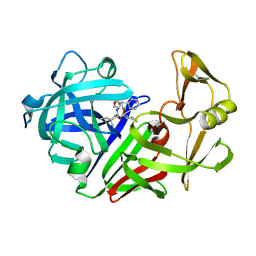 | |
5JIO
 
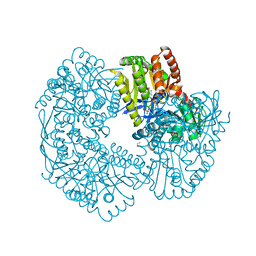 | | Structure of Mycobacterium thermoresistibile trehalose-6-phosphate synthase ternary complex with ADP and Glucose-6-phosphate. | | Descriptor: | 1,2-ETHANEDIOL, 6-O-phosphono-alpha-D-glucopyranose, ADENOSINE-5'-DIPHOSPHATE, ... | | Authors: | Mendes, V, Verma, N, Blaszczyk, M, Blundell, T.L. | | Deposit date: | 2016-04-22 | | Release date: | 2017-05-10 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.711 Å) | | Cite: | Mycobacterial OtsA Structures Unveil Substrate Preference Mechanism and Allosteric Regulation by 2-Oxoglutarate and 2-Phosphoglycerate.
Mbio, 10, 2019
|
|
5K5C
 
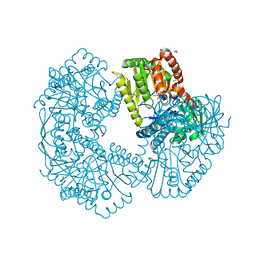 | | Structure of Mycobacterium thermoresistibile trehalose-6-phosphate synthase in a complex with Trehalose. | | Descriptor: | 1,2-ETHANEDIOL, 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, Alpha,alpha-trehalose-phosphate synthase, ... | | Authors: | Mendes, V, Verma, N, Blaszczyk, M, Blundell, T.L. | | Deposit date: | 2016-05-23 | | Release date: | 2017-06-07 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.848 Å) | | Cite: | Mycobacterial OtsA Structures Unveil Substrate Preference Mechanism and Allosteric Regulation by 2-Oxoglutarate and 2-Phosphoglycerate.
Mbio, 10, 2019
|
|
5K42
 
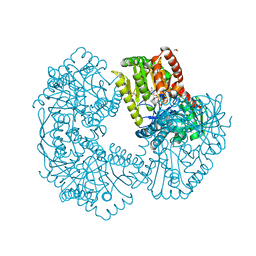 | | Structure of Mycobacterium thermoresistibile trehalose-6-phosphate synthase in a complex with GDP-glucose. | | Descriptor: | 1,2-ETHANEDIOL, 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, Alpha,alpha-trehalose-phosphate synthase, ... | | Authors: | Mendes, V, Verma, N, Blaszczyk, M, Blundell, T.L. | | Deposit date: | 2016-05-20 | | Release date: | 2017-06-21 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.921 Å) | | Cite: | Mycobacterial OtsA Structures Unveil Substrate Preference Mechanism and Allosteric Regulation by 2-Oxoglutarate and 2-Phosphoglycerate.
Mbio, 10, 2019
|
|
2WVI
 
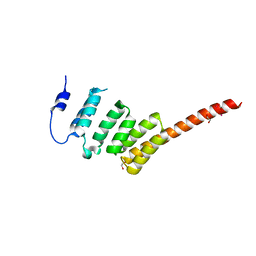 | | Crystal Structure of the N-terminal Domain of BubR1 | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, MITOTIC CHECKPOINT SERINE/THREONINE-PROTEIN KINASE BUB1 BETA, ... | | Authors: | D'Arcy, S, Davies, O.R, Blundell, T.L, Bolanos-Garcia, V.M. | | Deposit date: | 2009-10-16 | | Release date: | 2010-03-09 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Defining the Molecular Basis of Bubr1 Kinetochore Interactions and Anaphase-Promoting Complex/Cyclosome (Apc/C)-Cdc20 Inhibition
J.Biol.Chem., 285, 2010
|
|
1XY1
 
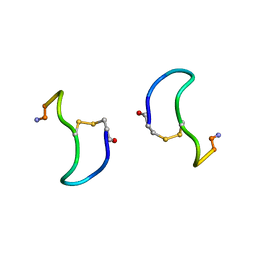 | | CRYSTAL STRUCTURE ANALYSIS OF DEAMINO-OXYTOCIN. CONFORMATIONAL FLEXIBILITY AND RECEPTOR BINDING | | Descriptor: | BETA-MERCAPTOPROPIONATE-OXYTOCIN | | Authors: | Husain, J, Blundell, T.L, Wood, S.P, Tickle, I.J, Cooper, S, Pitts, J.E. | | Deposit date: | 1987-06-05 | | Release date: | 1988-04-16 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.04 Å) | | Cite: | Crystal structure analysis of deamino-oxytocin: conformational flexibility and receptor binding.
Science, 232, 1986
|
|
1XY2
 
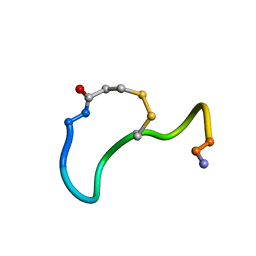 | | CRYSTAL STRUCTURE ANALYSIS OF DEAMINO-OXYTOCIN. CONFORMATIONAL FLEXIBILITY AND RECEPTOR BINDING | | Descriptor: | OXYTOCIN | | Authors: | Cooper, S, Blundell, T.L, Pitts, J.E, Wood, S.P, Tickle, I.J. | | Deposit date: | 1987-06-05 | | Release date: | 1988-04-16 | | Last modified: | 2025-03-26 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Crystal structure analysis of deamino-oxytocin: conformational flexibility and receptor binding.
Science, 232, 1986
|
|
4Y18
 
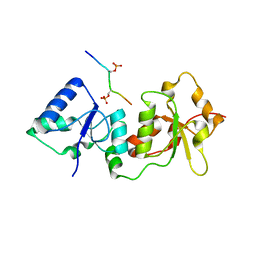 | |
4Y2G
 
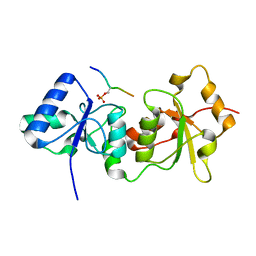 | |
4APE
 
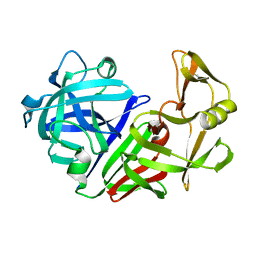 | | THE ACTIVE SITE OF ASPARTIC PROTEINASES | | Descriptor: | ENDOTHIAPEPSIN | | Authors: | Pearl, L.H, Sewell, B.T, Jenkins, J.A, Cooper, J.B, Blundell, T.L. | | Deposit date: | 1986-06-09 | | Release date: | 1986-07-14 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The Active Site of Aspartic Proteinases
FEBS Lett., 174, 1984
|
|
5CT2
 
 | | The structure of the NK1 fragment of HGF/SF complexed with CAPS | | Descriptor: | 3-CYCLOHEXYL-1-PROPYLSULFONIC ACID, Hepatocyte growth factor | | Authors: | Sigurdardottir, A.G, Winter, A, Sobkowicz, A, Fragai, M, Chirgadze, D.Y, Ascher, D.B, Blundell, T.L, Gherardi, E. | | Deposit date: | 2015-07-23 | | Release date: | 2015-08-12 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Exploring the chemical space of the lysine-binding pocket of the first kringle domain of hepatocyte growth factor/scatter factor (HGF/SF) yields a new class of inhibitors of HGF/SF-MET binding.
Chem Sci, 6, 2015
|
|
5CT3
 
 | | The structure of the NK1 fragment of HGF/SF complexed with 2FA | | Descriptor: | 3-hydroxypropane-1-sulfonic acid, Hepatocyte growth factor | | Authors: | Sigurdardottir, A.G, Winter, A, Sobkowicz, A, Fragai, M, Chirgadze, D.Y, Ascher, D.B, Blundell, T.L, Gherardi, E. | | Deposit date: | 2015-07-23 | | Release date: | 2015-08-12 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Exploring the chemical space of the lysine-binding pocket of the first kringle domain of hepatocyte growth factor/scatter factor (HGF/SF) yields a new class of inhibitors of HGF/SF-MET binding.
Chem Sci, 6, 2015
|
|
5CS3
 
 | | The structure of the NK1 fragment of HGF/SF complexed with (H)EPPS | | Descriptor: | 3-[4-(2-HYDROXYETHYL)PIPERAZIN-1-YL]PROPANE-1-SULFONIC ACID, Hepatocyte growth factor | | Authors: | Sigurdardottir, A.G, Winter, A, Sobkowicz, A, Fragai, M, Chirgadze, D.Y, Ascher, D.B, Blundell, T.L, Gherardi, E. | | Deposit date: | 2015-07-23 | | Release date: | 2015-08-12 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Exploring the chemical space of the lysine-binding pocket of the first kringle domain of hepatocyte growth factor/scatter factor (HGF/SF) yields a new class of inhibitors of HGF/SF-MET binding.
Chem Sci, 6, 2015
|
|
1ER8
 
 | | THE ACTIVE SITE OF ASPARTIC PROTEINASES | | Descriptor: | Endothiapepsin, H-77 | | Authors: | Hemmings, A.M, Veerapandian, B, Szelke, M, Cooper, J.B, Blundell, T.L. | | Deposit date: | 1989-10-16 | | Release date: | 1991-10-15 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The Active Site of Aspartic Proteinases
FEBS Lett., 174, 1984
|
|
6HXT
 
 | |
4CJM
 
 | | Crystal structure of human FGF18 | | Descriptor: | FIBROBLAST GROWTH FACTOR 18, SULFATE ION | | Authors: | Brown, A, Adam, L.E, Blundell, T.L. | | Deposit date: | 2013-12-21 | | Release date: | 2014-04-09 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | The Crystal Structure of Fibroblast Growth Factor 18 (Fgf18)
Protein Cell, 5, 2014
|
|
4D6P
 
 | | RADA C-TERMINAL ATPASE DOMAIN FROM PYROCOCCUS FURIOSUS BOUND TO AMPPNP | | Descriptor: | DNA REPAIR AND RECOMBINATION PROTEIN RADA, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Marsh, M.E, Ehebauer, M.T, Scott, D, Abell, C, Blundell, T.L, Hyvonen, M. | | Deposit date: | 2014-11-13 | | Release date: | 2015-01-14 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.482 Å) | | Cite: | ATP Half-Sites in Rada and Rad51 Recombinases Bind Nucleotides
FEBS Open Bio, 6, 2016
|
|
1XES
 
 | | Crystal structure of stilbene synthase from Pinus sylvestris | | Descriptor: | 3-(1H-INDOL-3-YL)-2-OXOPROPANOIC ACID, Dihydropinosylvin synthase | | Authors: | Ng, S.H, Chirgadze, D, Spiteller, D, Li, T.L, Spencer, J.B, Blundell, T.L. | | Deposit date: | 2004-09-12 | | Release date: | 2006-04-11 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of stilbene synthase from Pinus sylvestris
To be Published
|
|
