8JWZ
 
 | | Crystal structure of A2AR-T4L in complex with AB928 | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 3-[2-azanyl-6-[1-[[6-(2-oxidanylpropan-2-yl)pyridin-2-yl]methyl]-1,2,3-triazol-4-yl]pyrimidin-4-yl]-2-methyl-benzenecarbonitrile, Adenosine receptor A2a,Endolysin, ... | | Authors: | Weng, Y, Chen, Y, Xu, Y, Song, G. | | Deposit date: | 2023-06-29 | | Release date: | 2023-08-16 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.37 Å) | | Cite: | Structural insight into the dual-antagonistic mechanism of AB928 on adenosine A 2 receptors.
Sci China Life Sci, 67, 2024
|
|
8KGY
 
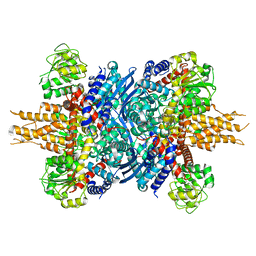 | | Human glutamate dehydrogenase I | | Descriptor: | Glutamate dehydrogenase 1, mitochondrial | | Authors: | Su, M.-Y. | | Deposit date: | 2023-08-20 | | Release date: | 2023-10-25 | | Last modified: | 2023-11-22 | | Method: | ELECTRON MICROSCOPY (2.59 Å) | | Cite: | Cryo-EM structure of the KLHL22 E3 ligase bound to an oligomeric metabolic enzyme.
Structure, 31, 2023
|
|
8KHP
 
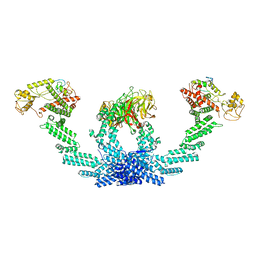 | | CULLIN3-KLHL22-RBX1 E3 ligase | | Descriptor: | Cullin-3, E3 ubiquitin-protein ligase RBX1, Kelch-like protein 22 | | Authors: | Su, M.-Y, Su, M.-Y. | | Deposit date: | 2023-08-22 | | Release date: | 2023-12-13 | | Method: | ELECTRON MICROSCOPY (3.67 Å) | | Cite: | Cryo-EM structure of the KLHL22 E3 ligase bound to an oligomeric metabolic enzyme.
Structure, 31, 2023
|
|
8K31
 
 | | The complex of WRKY33 C terminal DBD and SIB1 | | Descriptor: | Probable WRKY transcription factor 33, Sigma factor binding protein 1, chloroplastic, ... | | Authors: | Dong, X, Gong, Z, Hu, Y.F. | | Deposit date: | 2023-07-14 | | Release date: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Structural basis for the regulation of plant transcription factor WRKY33 by the VQ protein SIB1.
Commun Biol, 7, 2024
|
|
8IXB
 
 | | GMPCPP-Alpha1A/Beta2A-microtubule decorated with kinesin seam region | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, Kinesin-1 heavy chain, ... | | Authors: | Zheng, W, Zhao, Q.Y, Diao, L, Bao, L, Cong, Y. | | Deposit date: | 2023-03-31 | | Release date: | 2023-08-16 | | Last modified: | 2023-10-25 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Cryo-EM of alpha-tubulin isotype-containing microtubules revealed a contracted structure of alpha 4A/ beta 2A microtubules.
Acta Biochim.Biophys.Sin., 55, 2023
|
|
8IXE
 
 | | GMPCPP-Alpha1C/Beta2A-microtubule decorated with kinesin seam region | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, Kinesin-1 heavy chain, ... | | Authors: | Zheng, W, Zhao, Q.Y, Diao, L, Bao, L, Cong, Y. | | Deposit date: | 2023-03-31 | | Release date: | 2023-08-16 | | Last modified: | 2023-10-25 | | Method: | ELECTRON MICROSCOPY (4.4 Å) | | Cite: | Cryo-EM of alpha-tubulin isotype-containing microtubules revealed a contracted structure of alpha 4A/ beta 2A microtubules.
Acta Biochim.Biophys.Sin., 55, 2023
|
|
8IXG
 
 | | GMPCPP-Alpha4A/Beta2A-microtubule decorated with kinesin seam region | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, Kinesin-1 heavy chain, ... | | Authors: | Zheng, W, Zhao, Q.Y, Diao, L, Bao, L, Cong, Y. | | Deposit date: | 2023-03-31 | | Release date: | 2023-08-16 | | Last modified: | 2023-10-25 | | Method: | ELECTRON MICROSCOPY (4.4 Å) | | Cite: | Cryo-EM of alpha-tubulin isotype-containing microtubules revealed a contracted structure of alpha 4A/ beta 2A microtubules.
Acta Biochim.Biophys.Sin., 55, 2023
|
|
8IXD
 
 | | GMPCPP-Alpha1C/Beta2A-microtubule decorated with kinesin non-seam region | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, Kinesin-1 heavy chain, ... | | Authors: | Zheng, W, Zhao, Q.Y, Diao, L, Bao, L, Cong, Y. | | Deposit date: | 2023-03-31 | | Release date: | 2023-08-16 | | Last modified: | 2023-10-25 | | Method: | ELECTRON MICROSCOPY (4.4 Å) | | Cite: | Cryo-EM of alpha-tubulin isotype-containing microtubules revealed a contracted structure of alpha 4A/ beta 2A microtubules.
Acta Biochim.Biophys.Sin., 55, 2023
|
|
8IXA
 
 | | GMPCPP-Alpha1A/Beta2A-microtubule decorated with kinesin non-seam region | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, Kinesin-1 heavy chain, ... | | Authors: | Zheng, W, Zhao, Q.Y, Diao, L, Bao, L, Cong, Y. | | Deposit date: | 2023-03-31 | | Release date: | 2023-08-16 | | Last modified: | 2023-10-25 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Cryo-EM of alpha-tubulin isotype-containing microtubules revealed a contracted structure of alpha 4A/ beta 2A microtubules.
Acta Biochim.Biophys.Sin., 55, 2023
|
|
8IXF
 
 | | GMPCPP-Alpha4A/Beta2A-microtubule decorated with kinesin non-seam region | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, Kinesin-1 heavy chain, ... | | Authors: | Zheng, W, Zhao, Q.Y, Diao, L, Bao, L, Cong, Y. | | Deposit date: | 2023-03-31 | | Release date: | 2023-08-16 | | Last modified: | 2023-10-25 | | Method: | ELECTRON MICROSCOPY (4.4 Å) | | Cite: | Cryo-EM of alpha-tubulin isotype-containing microtubules revealed a contracted structure of alpha 4A/ beta 2A microtubules.
Acta Biochim.Biophys.Sin., 55, 2023
|
|
4ER1
 
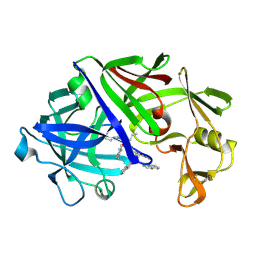 | | THE ACTIVE SITE OF ASPARTIC PROTEINASES | | Descriptor: | ENDOTHIAPEPSIN, N-[(1R,2R,4R)-1-(cyclohexylmethyl)-2-hydroxy-6-methyl-4-{[(2R)-2-methylbutyl]carbamoyl}heptyl]-3-(1H-imidazol-3-ium-4-yl)-N~2~-[3-naphthalen-1-yl-2-(naphthalen-1-ylmethyl)propanoyl]-L-alaninamide | | Authors: | Quail, J.W, Cooper, J.B, Szelke, M, Blundell, T.L. | | Deposit date: | 1990-10-14 | | Release date: | 1991-01-15 | | Last modified: | 2017-11-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The active site of aspartic proteinases
FEBS Lett., 174, 1984
|
|
6IYN
 
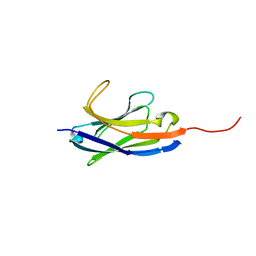 | | Solution structure of camelid nanobody Nb26 against aflatoxin B1 | | Descriptor: | Nb26 | | Authors: | Nie, Y, He, T, Zhu, J, Li, S.L, Hu, R, Yang, Y.H. | | Deposit date: | 2018-12-17 | | Release date: | 2019-01-23 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Chemical shift assignments of a camelid nanobody against aflatoxin B1.
Biomol NMR Assign, 13, 2019
|
|
7WFC
 
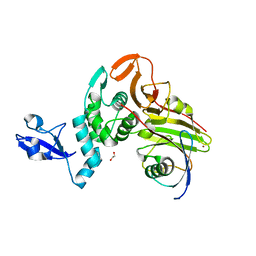 | | X-ray structure of HKU1-PLP2(Cys109Ser) catalytic mutant in complex with free ubiquitin | | Descriptor: | 1,2-ETHANEDIOL, 60S ribosomal protein L40, Papain-like protease, ... | | Authors: | Xiong, Y.X, Fu, Z.Y, Huang, H. | | Deposit date: | 2021-12-26 | | Release date: | 2022-12-28 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The substrate selectivity of papain-like proteases from human-infecting coronaviruses correlates with innate immune suppression.
Sci.Signal., 16, 2023
|
|
2ER7
 
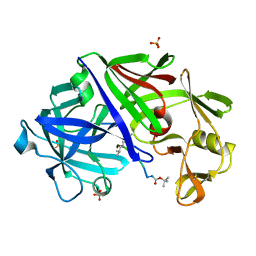 | | X-RAY ANALYSES OF ASPARTIC PROTEINASES.III. THREE-DIMENSIONAL STRUCTURE OF ENDOTHIAPEPSIN COMPLEXED WITH A TRANSITION-STATE ISOSTERE INHIBITOR OF RENIN AT 1.6 ANGSTROMS RESOLUTION | | Descriptor: | ENDOTHIAPEPSIN, SULFATE ION, TRANSITION-STATE ISOSTERE INHIBITOR OF RENIN | | Authors: | Veerapandian, B, Cooper, J.B, Szelke, M, Blundell, T.L. | | Deposit date: | 1990-11-12 | | Release date: | 1991-01-15 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | X-ray analyses of aspartic proteinases. III Three-dimensional structure of endothiapepsin complexed with a transition-state isostere inhibitor of renin at 1.6 A resolution.
J.Mol.Biol., 216, 1990
|
|
6LRM
 
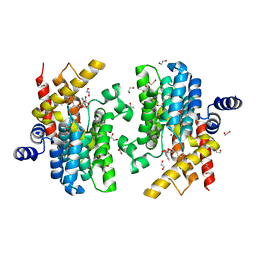 | | Crystal structure of PDE4D catalytic domain in complex with arctigenin | | Descriptor: | 1,2-ETHANEDIOL, Arctigenin, MAGNESIUM ION, ... | | Authors: | Zhang, X.L, Li, M.J, Xu, Y.C. | | Deposit date: | 2020-01-16 | | Release date: | 2021-04-14 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Identification of phosphodiesterase-4 as the therapeutic target of arctigenin in alleviating psoriatic skin inflammation.
J Adv Res, 33, 2021
|
|
6LK8
 
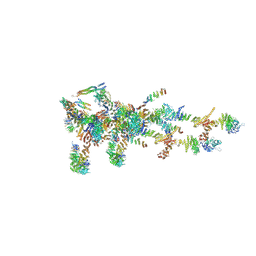 | | Structure of Xenopus laevis Cytoplasmic Ring subunit. | | Descriptor: | GATOR complex protein SEC13, MGC154553 protein, MGC83295 protein, ... | | Authors: | Shi, Y, Huang, G, Yan, C, Zhang, Y. | | Deposit date: | 2019-12-18 | | Release date: | 2021-07-21 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (5.5 Å) | | Cite: | Structure of the cytoplasmic ring of the Xenopus laevis nuclear pore complex by cryo-electron microscopy single particle analysis.
Cell Res., 30, 2020
|
|
6LMR
 
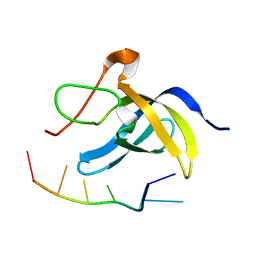 | | Solution structure of cold shock domain and ssDNA complex | | Descriptor: | DNA (5'-D(P*AP*AP*CP*AP*CP*CP*T)-3'), Y-box-binding protein 1 | | Authors: | Fan, J, Yang, D. | | Deposit date: | 2019-12-26 | | Release date: | 2020-07-22 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural basis of DNA binding to human YB-1 cold shock domain regulated by phosphorylation.
Nucleic Acids Res., 48, 2020
|
|
6K3H
 
 | |
6LMS
 
 | |
2BKJ
 
 | |
2IRW
 
 | | Human 11-beta-Hydroxysteroid Dehydrogenase (HSD1) with NADP and Adamantane Ether Inhibitor | | Descriptor: | (1S,3R,4S,5S,7S)-4-{[2-(4-METHOXYPHENOXY)-2-METHYLPROPANOYL]AMINO}ADAMANTANE-1-CARBOXAMIDE, Corticosteroid 11-beta-dehydrogenase isozyme 1, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Longenecker, K.L, Patel, J.R, Russell, J, Qin, W. | | Deposit date: | 2006-10-16 | | Release date: | 2007-01-23 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Discovery of adamantane ethers as inhibitors of 11beta-HSD-1: Synthesis and biological evaluation.
Bioorg.Med.Chem.Lett., 17, 2007
|
|
7DVM
 
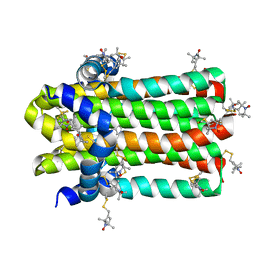 | | DgkA structure in E.coli lipid bilayer | | Descriptor: | Diacylglycerol kinase | | Authors: | Li, J, Yang, J. | | Deposit date: | 2021-01-13 | | Release date: | 2022-04-13 | | Last modified: | 2023-09-27 | | Method: | SOLID-STATE NMR | | Cite: | Structure of membrane diacylglycerol kinase in lipid bilayers.
Commun Biol, 4, 2021
|
|
7FIL
 
 | |
7C2X
 
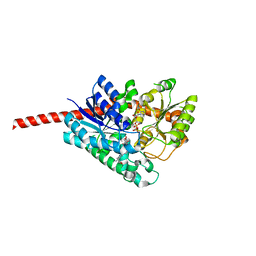 | | Crystal Structure of Glycyrrhiza uralensis UGT73P12 complexed with glycyrrhetinic acid 3-O-monoglucuronide | | Descriptor: | (2~{S},3~{S},4~{S},5~{R},6~{R})-6-[[(3~{S},4~{a}~{R},6~{a}~{R},6~{b}~{S},8~{a}~{S},11~{S},12~{a}~{R},14~{a}~{R},14~{b}~{S})-11-carboxy-4,4,6~{a},6~{b},8~{a},11,14~{b}-heptamethyl-14-oxidanylidene-2,3,4~{a},5,6,7,8,9,10,12,12~{a},14~{a}-dodecahydro-1~{H}-picen-3-yl]oxy]-3,4,5-tris(oxidanyl)oxane-2-carboxylic acid, Glycosyltransferase, URIDINE-5'-DIPHOSPHATE | | Authors: | Ren, J. | | Deposit date: | 2020-05-09 | | Release date: | 2021-05-12 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Crystal Structure of Glycyrrhiza uralensis UGT73P12 complexed with glycyrrhetinic acid 3-O-monoglucuronide
To Be Published
|
|
7FIK
 
 | | The cryo-EM structure of the CR subunit from X. laevis NPC | | Descriptor: | MGC154553 protein, MGC83295 protein, MGC83926 protein, ... | | Authors: | Shi, Y, Huang, G, Zhan, X. | | Deposit date: | 2021-07-31 | | Release date: | 2022-11-09 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Structure of the cytoplasmic ring of the Xenopus laevis nuclear pore complex.
Science, 376, 2022
|
|
