7VGL
 
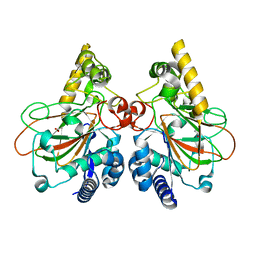 | | Crystal structure of CmnC | | Descriptor: | ACETATE ION, CmnC | | Authors: | Hsiao, Y.H, Huang, S.J, Lin, E.C, Lee, Y.C, Zheng, Y.Z, Chang, C.Y. | | Deposit date: | 2021-09-17 | | Release date: | 2022-09-21 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Crystal structure of the alpha-ketoglutarate-dependent non-heme iron oxygenase CmnC in capreomycin biosynthesis and its engineering to catalyze hydroxylation of the substrate enantiomer.
Front Chem, 10, 2022
|
|
7WBQ
 
 | |
7WBP
 
 | |
7WBL
 
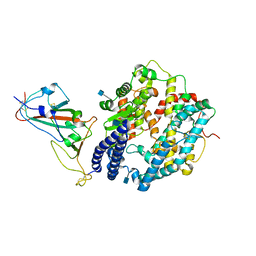 | | Cryo-EM structure of human ACE2 complexed with SARS-CoV-2 Omicron RBD | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, Spike protein S1, ... | | Authors: | Liu, S, Gao, F.G. | | Deposit date: | 2021-12-17 | | Release date: | 2022-01-19 | | Last modified: | 2022-03-02 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Receptor binding and complex structures of human ACE2 to spike RBD from omicron and delta SARS-CoV-2.
Cell, 185, 2022
|
|
7W0W
 
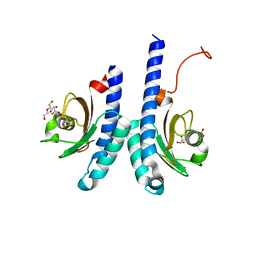 | | The novel membrane-proximal sensing mechanism in a broad-ligand binding chemoreceptor McpA of Bacillus velezensis | | Descriptor: | (2S)-2-hydroxybutanedioic acid, 1,2-ETHANEDIOL, ACETATE ION, ... | | Authors: | Feng, H.C, Shen, Q.R, Zhang, R.F. | | Deposit date: | 2021-11-18 | | Release date: | 2022-07-06 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.247 Å) | | Cite: | Signal binding at both modules of its dCache domain enables the McpA chemoreceptor of Bacillus velezensis to sense different ligands.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7WSE
 
 | |
7WSF
 
 | |
7WEG
 
 | | Complex structure of PDZD7 and FCHSD2 | | Descriptor: | FCHSD2, PDZ domain-containing protein 7, ZINC ION | | Authors: | Wang, H, Lin, L, Lu, Q. | | Deposit date: | 2021-12-23 | | Release date: | 2022-11-16 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Deafness-related protein PDZD7 forms complex with the C-terminal tail of FCHSD2.
Biochem.J., 479, 2022
|
|
7WSH
 
 | | Cryo-EM structure of SARS-CoV-2 spike receptor-binding domain in complex with sea lion ACE2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme, Spike protein S1, ... | | Authors: | Li, S, Han, P, Qi, J. | | Deposit date: | 2022-01-29 | | Release date: | 2022-11-09 | | Method: | ELECTRON MICROSCOPY (2.89 Å) | | Cite: | Cross-species recognition and molecular basis of SARS-CoV-2 and SARS-CoV binding to ACE2s of marine animals.
Natl Sci Rev, 9, 2022
|
|
7WSG
 
 | |
7XA7
 
 | | Crystal structure of SARS-CoV-2 receptor-binding domain in complex with intermediate horseshoe bat ACE2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme, Spike protein S1, ... | | Authors: | Tang, L.F, Zhang, D, Han, P, Qi, J.X. | | Deposit date: | 2022-03-17 | | Release date: | 2022-12-21 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.31 Å) | | Cite: | Structural basis of SARS-CoV-2 and its variants binding to intermediate horseshoe bat ACE2.
Int J Biol Sci, 18, 2022
|
|
7Y5F
 
 | | Crystal structure of CmnC in complex with L-homoarginine | | Descriptor: | CmnC, FE (III) ION, L(+)-TARTARIC ACID, ... | | Authors: | Hsiao, Y.H, Huang, S.J, Lin, E.C, Lee, Y.C, Chang, C.Y. | | Deposit date: | 2022-06-17 | | Release date: | 2023-07-05 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | Crystal structure of the alpha-ketoglutarate-dependent non-heme iron oxygenase CmnC in capreomycin biosynthesis and its engineering to catalyze hydroxylation of the substrate enantiomer.
Front Chem, 10, 2022
|
|
7Y5I
 
 | | Crystal structure of CmnC in complex with L-homoarginine | | Descriptor: | ARGININE, CmnC, FE (III) ION, ... | | Authors: | Hsiao, Y.H, Huang, S.J, Lin, E.C, Lee, Y.C, Chang, C.Y. | | Deposit date: | 2022-06-17 | | Release date: | 2023-07-05 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Crystal structure of the alpha-ketoglutarate-dependent non-heme iron oxygenase CmnC in capreomycin biosynthesis and its engineering to catalyze hydroxylation of the substrate enantiomer.
Front Chem, 10, 2022
|
|
7Y5P
 
 | | Crystal structure of CmnC in complex with L-arginine and alpha-KG | | Descriptor: | 2-OXOGLUTARIC ACID, ARGININE, CmnC, ... | | Authors: | Hsiao, Y.H, Huang, S.J, Lin, E.C, Lee, Y.C, Chang, C.Y. | | Deposit date: | 2022-06-17 | | Release date: | 2023-07-05 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of the alpha-ketoglutarate-dependent non-heme iron oxygenase CmnC in capreomycin biosynthesis and its engineering to catalyze hydroxylation of the substrate enantiomer.
Front Chem, 10, 2022
|
|
7YHE
 
 | | Crystal structure of the triple mutant CmnC-L136Q,S138G,D249Y in complex with alpha-KG | | Descriptor: | 2-OXOGLUTARIC ACID, CmnC, FE (III) ION, ... | | Authors: | Huang, S.J, Hsiao, Y.H, Lin, E.C, Hsiao, P.Y, Chang, C.Y. | | Deposit date: | 2022-07-13 | | Release date: | 2023-07-19 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Crystal structure of the alpha-ketoglutarate-dependent non-heme iron oxygenase CmnC in capreomycin biosynthesis and its engineering to catalyze hydroxylation of the substrate enantiomer.
Front Chem, 10, 2022
|
|
3TPZ
 
 | |
7WRI
 
 | | Cryo-EM structure of SARS-CoV-2 Omicron spike receptor-binding domain in complex with mouse ACE2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Processed angiotensin-converting enzyme 2, Spike glycoprotein, ... | | Authors: | Han, P, Xie, Y, Qi, J. | | Deposit date: | 2022-01-26 | | Release date: | 2022-06-08 | | Last modified: | 2022-12-21 | | Method: | ELECTRON MICROSCOPY (3.03 Å) | | Cite: | Broader-species receptor binding and structural bases of Omicron SARS-CoV-2 to both mouse and palm-civet ACE2s.
Cell Discov, 8, 2022
|
|
7WSK
 
 | | Crystal structure of SARS-CoV-2 Omicron spike receptor-binding domain in complex with civet ACE2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Processed angiotensin-converting enzyme 2, Spike protein S1, ... | | Authors: | Huang, B, Han, P, Qi, J. | | Deposit date: | 2022-01-29 | | Release date: | 2022-06-08 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Broader-species receptor binding and structural bases of Omicron SARS-CoV-2 to both mouse and palm-civet ACE2s.
Cell Discov, 8, 2022
|
|
7WRH
 
 | | Cryo-EM structure of SARS-CoV-2 Omicron BA.1 spike protein in complex with mouse ACE2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Processed angiotensin-converting enzyme 2, ... | | Authors: | Han, P, Xie, Y, Qi, J. | | Deposit date: | 2022-01-26 | | Release date: | 2023-02-01 | | Method: | ELECTRON MICROSCOPY (2.66 Å) | | Cite: | Broader-species receptor binding and structural bases of Omicron SARS-CoV-2 to both mouse and palm-civet ACE2s.
Cell Discov, 8, 2022
|
|
1AEL
 
 | |
6MN2
 
 | | Crystal structure of meta-AAC0038, an environmental aminoglycoside resistance enzyme, mutant H168A in abortive complex with sisomicin-CoA | | Descriptor: | (1S,2S,3R,4S,6R)-4,6-diamino-3-{[(2S,3R)-3-amino-6-(aminomethyl)-3,4-dihydro-2H-pyran-2-yl]oxy}-2-hydroxycyclohexyl 3-deoxy-4-C-methyl-3-(methylamino)-beta-L-arabinopyranoside, Aminoglycoside N(3)-acetyltransferase, CHLORIDE ION, ... | | Authors: | Stogios, P.J, Skarina, T, Michalska, K, Xu, Z, Yim, V, Savchenko, A, Joachimiak, A, Satchell, K.J, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2018-10-01 | | Release date: | 2018-10-24 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.744 Å) | | Cite: | Crystal structure of meta-AAC0038, an environmental aminoglycoside resistance enzyme, mutant H168A in abortive complex with sisomicin-CoA
To Be Published
|
|
6MN1
 
 | | Crystal structure of meta-AAC0038, an environmental aminoglycoside resistance enzyme, mutant H168A in abortive complex with gentamicin-CoA | | Descriptor: | (2R,3R,4R,5R)-2-((1S,2S,3R,4S,6R)-4,6-DIAMINO-3-((2R,3R,6S)-3-AMINO-6-(AMINOMETHYL)-TETRAHYDRO-2H-PYRAN-2-YLOXY)-2-HYDR OXYCYCLOHEXYLOXY)-5-METHYL-4-(METHYLAMINO)-TETRAHYDRO-2H-PYRAN-3,5-DIOL, 3,6,9,12,15,18,21,24,27,30,33,36,39-TRIDECAOXAHENTETRACONTANE-1,41-DIOL, Aminoglycoside N(3)-acetyltransferase, ... | | Authors: | Stogios, P.J, Skarina, T, Michalska, K, Xu, Z, Yim, V, Savchenko, A, Joachimiak, A, Satchell, K.J, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2018-10-01 | | Release date: | 2018-10-24 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Crystal structure of meta-AAC0038, an environmental aminoglycoside resistance enzyme, mutant H168A in abortive complex with gentamicin-CoA
To Be Published
|
|
1B4M
 
 | | NMR STRUCTURE OF APO CELLULAR RETINOL-BINDING PROTEIN II, 24 STRUCTURES | | Descriptor: | CELLULAR RETINOL-BINDING PROTEIN II | | Authors: | Lu, J, Lin, C.-L, Tang, C, Ponder, J.W, Kao, J.L.F, Cistola, D.P, Li, E. | | Deposit date: | 1998-12-23 | | Release date: | 1999-04-27 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | The structure and dynamics of rat apo-cellular retinol-binding protein II in solution: comparison with the X-ray structure.
J.Mol.Biol., 286, 1999
|
|
