7QVM
 
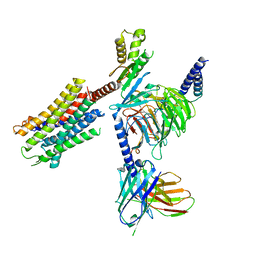 | | Human Oxytocin receptor (OTR) oxytocin Gq chimera (mGoqi) complex | | Descriptor: | Antibody fragment scFv16, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Waltenspuhl, Y, Ehrenmann, J, Vacca, S, Thom, C, Medalia, O, Pluckthun, A. | | Deposit date: | 2022-01-21 | | Release date: | 2022-08-10 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.25 Å) | | Cite: | Structural basis for the activation and ligand recognition of the human oxytocin receptor.
Nat Commun, 13, 2022
|
|
8RK0
 
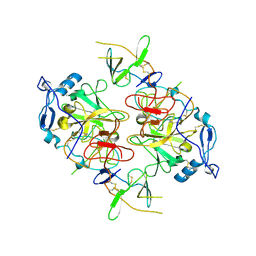 | | HCV E1/E2 homodimer complex, ectodomain | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, HCV E1, ... | | Authors: | Augestad, E.H, Olesen, C.H, Groenberg, C, Soerensen, A, Velazquez-Moctezuma, R, Fanalista, M, Bukh, J, Wang, K, Gourdon, P, Prentoe, J. | | Deposit date: | 2023-12-22 | | Release date: | 2024-09-04 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.38 Å) | | Cite: | The hepatitis C virus envelope protein complex is a dimer of heterodimers.
Nature, 633, 2024
|
|
8RJJ
 
 | | HCV E1/E2 homodimer complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Genome polyprotein, ... | | Authors: | Augestad, E.H, Olesen, C.H, Groenberg, C, Soerensen, A, Velazquez-Moctezuma, R, Fanalista, M, Bukh, J, Wang, K, Gourdon, P, Prentoe, J. | | Deposit date: | 2023-12-21 | | Release date: | 2024-09-04 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.55 Å) | | Cite: | The hepatitis C virus envelope protein complex is a dimer of heterodimers.
Nature, 633, 2024
|
|
8OTO
 
 | |
8OSX
 
 | |
8OTR
 
 | | SARS-CoV-2 nsp10-16 methyltransferase in complex with SAM analog BDH 33959089 | | Descriptor: | (2S,3S,4R,5R)-5-(6-aminopurin-9-yl)-N-(1-methylpiperidin-4-yl)-3,4-bis(oxidanyl)oxolane-2-carboxamide, 1,2-ETHANEDIOL, 2'-O-methyltransferase nsp16, ... | | Authors: | Kremling, V, Sprenger, J, Oberthuer, D. | | Deposit date: | 2023-04-21 | | Release date: | 2023-05-24 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Crystal structures of Tubercidin and Adenosine bound to the active site of the SARS-CoV-2 methyltransferase nsp10-16
To Be Published
|
|
6Z2Q
 
 | | Crystal structure of wild type OgpA from Akkermansia muciniphila in complex with an O-glycopeptide (GalGalNAc-TS) product | | Descriptor: | 1,2-ETHANEDIOL, ACETIC ACID, Glycodrosocin, ... | | Authors: | Trastoy, B, Naegali, A, Anso, I, Sjogren, J, Guerin, M.E. | | Deposit date: | 2020-05-18 | | Release date: | 2020-09-30 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.347 Å) | | Cite: | Structural basis of mammalian mucin processing by the human gut O-glycopeptidase OgpA from Akkermansia muciniphila.
Nat Commun, 11, 2020
|
|
8OV4
 
 | |
8OV1
 
 | |
8OV3
 
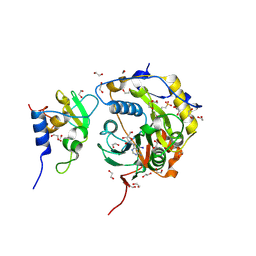 | | SARS-CoV-2 nsp10-16 methyltransferase in complex with 5-Iodotubercidin | | Descriptor: | (2R,3R,4S,5R)-2-(4-AMINO-5-IODO-7H-PYRROLO[2,3-D]PYRIMIDIN-7-YL)-5-(HYDROXYMETHYL)TETRAHYDROFURAN-3,4-DIOL, 1,2-ETHANEDIOL, 2'-O-methyltransferase nsp16, ... | | Authors: | Kremling, V, Sprenger, J, Oberthuer, D. | | Deposit date: | 2023-04-25 | | Release date: | 2023-05-10 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Crystal structures of Tubercidin and Adenosine bound to the active site of the SARS-CoV-2 methyltransferase nsp10-16
To Be Published
|
|
8OT0
 
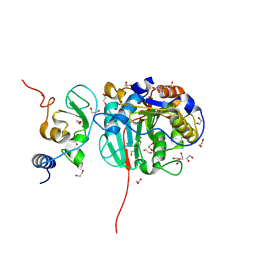 | | SARS-CoV-2 nsp10-16 methyltransferase in complex with MTA and glycine | | Descriptor: | 1,2-ETHANEDIOL, 2'-O-methyltransferase nsp16, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, ... | | Authors: | Kremling, V, Sprenger, J, Oberthuer, D, Falke, S. | | Deposit date: | 2023-04-20 | | Release date: | 2023-05-10 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Crystal structures of Tubercidin and Adenosine bound to the active site of the SARS-CoV-2 methyltransferase nsp10-16
To Be Published
|
|
8OV2
 
 | |
6Z2D
 
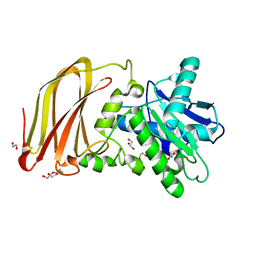 | | Crystal structure of wild type OgpA from Akkermansia muciniphila in P 41 21 2 | | Descriptor: | 1,2-ETHANEDIOL, FORMIC ACID, O-glycan protease, ... | | Authors: | Trastoy, B, Naegali, A, Anso, I, Sjogren, J, Guerin, M.E. | | Deposit date: | 2020-05-15 | | Release date: | 2020-09-30 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.899 Å) | | Cite: | Structural basis of mammalian mucin processing by the human gut O-glycopeptidase OgpA from Akkermansia muciniphila.
Nat Commun, 11, 2020
|
|
6Z2O
 
 | | Crystal structure of wild type OgpA from Akkermansia muciniphila in P 21 21 21 | | Descriptor: | 1,2-ETHANEDIOL, O-glycan protease, ZINC ION | | Authors: | Trastoy, B, Naegali, A, Anso, I, Sjogren, J, Guerin, M.E. | | Deposit date: | 2020-05-18 | | Release date: | 2020-09-30 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.649 Å) | | Cite: | Structural basis of mammalian mucin processing by the human gut O-glycopeptidase OgpA from Akkermansia muciniphila.
Nat Commun, 11, 2020
|
|
6Z2P
 
 | | Crystal structure of catalytic inactive OgpA from Akkermansia muciniphila in complex with an O-glycopeptide (glycodrosocin) substrate | | Descriptor: | CALCIUM ION, Glycodrosocin, O-glycan protease, ... | | Authors: | Trastoy, B, Naegali, A, Anso, I, Sjogren, J, Guerin, M.E. | | Deposit date: | 2020-05-18 | | Release date: | 2020-09-30 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.16 Å) | | Cite: | Structural basis of mammalian mucin processing by the human gut O-glycopeptidase OgpA from Akkermansia muciniphila.
Nat Commun, 11, 2020
|
|
4ETC
 
 | | Lysozyme, room temperature, 24 kGy dose | | Descriptor: | CHLORIDE ION, Lysozyme C, SODIUM ION | | Authors: | Boutet, S, Lomb, L, Williams, G, Barends, T, Aquila, A, Doak, R.B, Weierstall, U, DePonte, D, Steinbrener, J, Shoeman, R, Messerschmidt, M, Barty, A, White, T, Kassemeyer, S, Kirian, R, Seibert, M, Montanez, P, Kenney, C, Herbst, R, Hart, P, Pines, J, Haller, G, Gruner, S, Philllip, H, Tate, M, Hromalik, M, Koerner, L, van Bakel, N, Morse, J, Ghonsalves, W, Arnlund, D, Bogan, M, Calemann, C, Fromme, R, Hampton, C, Hunter, M, Johansson, L, Katona, G, Kupitz, C, Liang, M, Martin, A, Nass, K, Redecke, L, Stellato, F, Timneanu, N, Wang, D, Zatsepin, N, Schafer, D, Defever, K, Neutze, R, Fromme, P, Spence, J, Chapman, H, Schlichting, I. | | Deposit date: | 2012-04-24 | | Release date: | 2012-06-13 | | Last modified: | 2017-11-15 | | Method: | X-RAY DIFFRACTION (1.906 Å) | | Cite: | High-resolution protein structure determination by serial femtosecond crystallography.
Science, 337, 2012
|
|
4ETA
 
 | | Lysozyme, room temperature, 400 kGy dose | | Descriptor: | CHLORIDE ION, Lysozyme C | | Authors: | Boutet, S, Lomb, L, Williams, G, Barends, T, Aquila, A, Doak, R.B, Weierstall, U, DePonte, D, Steinbrener, J, Shoeman, R, Messerschmidt, M, Barty, A, White, T, Kassemeyer, S, Kirian, R, Seibert, M, Montanez, P, Kenney, C, Herbst, R, Hart, P, Pines, J, Haller, G, Gruner, S, Philllip, H, Tate, M, Hromalik, M, Koerner, L, van Bakel, N, Morse, J, Ghonsalves, W, Arnlund, D, Bogan, M, Calemann, C, Fromme, R, Hampton, C, Hunter, M, Johansson, L, Katona, G, Kupitz, C, Liang, M, Martin, A, Nass, K, Redecke, L, Stellato, F, Timneanu, N, Wang, D, Zatsepin, N, Schafer, D, Defever, K, Neutze, R, Fromme, P, Spence, J, Chapman, H, Schlichting, I. | | Deposit date: | 2012-04-24 | | Release date: | 2012-06-13 | | Last modified: | 2017-11-15 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | High-resolution protein structure determination by serial femtosecond crystallography.
Science, 337, 2012
|
|
4ETB
 
 | | lysozyme, room temperature, 200 kGy dose | | Descriptor: | CHLORIDE ION, Lysozyme C | | Authors: | Boutet, S, Lomb, L, Williams, G, Barends, T, Aquila, A, Doak, R.B, Weierstall, U, DePonte, D, Steinbrener, J, Shoeman, R, Messerschmidt, M, Barty, A, White, T, Kassemeyer, S, Kirian, R, Seibert, M, Montanez, P, Kenney, C, Herbst, R, Hart, P, Pines, J, Haller, G, Gruner, S, Philllip, H, Tate, M, Hromalik, M, Koerner, L, van Bakel, N, Morse, J, Ghonsalves, W, Arnlund, D, Bogan, M, Calemann, C, Fromme, R, Hampton, C, Hunter, M, Johansson, L, Katona, G, Kupitz, C, Liang, M, Martin, A, Nass, K, Redecke, L, Stellato, F, Timneanu, N, Wang, D, Zatsepin, N, Schafer, D, Defever, K, Neutze, R, Fromme, P, Spence, J, Chapman, H, Schlichting, I. | | Deposit date: | 2012-04-24 | | Release date: | 2012-06-13 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.908 Å) | | Cite: | High-resolution protein structure determination by serial femtosecond crystallography.
Science, 337, 2012
|
|
4ET8
 
 | | Hen egg-white lysozyme solved from 40 fs free-electron laser pulse data | | Descriptor: | CHLORIDE ION, Lysozyme C, SODIUM ION | | Authors: | Boutet, S, Lomb, L, Williams, G, Barends, T, Aquila, A, Doak, R.B, Weierstall, U, DePonte, D, Steinbrener, J, Shoeman, R, Messerschmidt, M, Barty, A, White, T, Kassemeyer, S, Kirian, R, Seibert, M, Montanez, P, Kenney, C, Herbst, R, Hart, P, Pines, J, Haller, G, Gruner, S, Philllip, H, Tate, M, Hromalik, M, Koerner, L, van Bakel, N, Morse, J, Ghonsalves, W, Arnlund, D, Bogan, M, Calemann, C, Fromme, R, Hampton, C, Hunter, M, Johansson, L, Katona, G, Kupitz, C, Liang, M, Martin, A, Nass, K, Redecke, L, Stellato, F, Timneanu, N, Wang, D, Zatsepin, N, Schafer, D, Defever, K, Neutze, R, Fromme, P, Spence, J, Chapman, H, Schlichting, I. | | Deposit date: | 2012-04-24 | | Release date: | 2012-06-13 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | High-resolution protein structure determination by serial femtosecond crystallography.
Science, 337, 2012
|
|
4ETE
 
 | | Lysozyme, room-temperature, rotating anode, 0.0021 MGy | | Descriptor: | CHLORIDE ION, Lysozyme C | | Authors: | Boutet, S, Lomb, L, Williams, G, Barends, T, Aquila, A, Doak, R.B, Weierstall, U, DePonte, D, Steinbrener, J, Shoeman, R, Messerschmidt, M, Barty, A, White, T, Kassemeyer, S, Kirian, R, Seibert, M, Montanez, P, Kenney, C, Herbst, R, Hart, P, Pines, J, Haller, G, Gruner, S, Philllip, H, Tate, M, Hromalik, M, Koerner, L, van Bakel, N, Morse, J, Ghonsalves, W, Arnlund, D, Bogan, M, Calemann, C, Fromme, R, Hampton, C, Hunter, M, Johansson, L, Katona, G, Kupitz, C, Liang, M, Martin, A, Nass, K, Redecke, L, Stellato, F, Timneanu, N, Wang, D, Zatsepin, N, Schafer, D, Defever, K, Neutze, R, Fromme, P, Spence, J, Chapman, H, Schlichting, I. | | Deposit date: | 2012-04-24 | | Release date: | 2012-06-13 | | Last modified: | 2017-11-15 | | Method: | X-RAY DIFFRACTION (1.905 Å) | | Cite: | High-resolution protein structure determination by serial femtosecond crystallography.
Science, 337, 2012
|
|
4CAS
 
 | | Serial femtosecond crystallography structure of a photosynthetic reaction center | | Descriptor: | (2E,6E,10E,14E,18E,22E,26E)-3,7,11,15,19,23,27,31-OCTAMETHYLDOTRIACONTA-2,6,10,14,18,22,26,30-OCTAENYL TRIHYDROGEN DIPHOSPHATE, 15-cis-1,2-dihydroneurosporene, BACTERIOCHLOROPHYLL A, ... | | Authors: | Johansson, L.C, Arnlund, D, Katona, G, White, T.A, Barty, A, DePonte, D.P, Shoeman, R.L, Wickstrand, C, Sharma, A, Williams, G.J, Aquila, A, Bogan, M.J, Caleman, C, Davidsson, J, Doak, R.B, Frank, M, Fromme, R, Galli, L, Grotjohann, I, Hunter, M.S, Kassemeyer, S, Kirian, R.A, Kupitz, C, Liang, M, Lomb, L, Malmerberg, E, Martin, A.V, Messerschmidt, M, Nass, K, Redecke, L, Seibert, M.M, Sjohamn, J, Steinbrener, J, Stellato, F, Wang, D, Wahlgren, W.Y, Weierstall, U, Westenhoff, S, Zatsepin, N.A, Boutet, S, Spence, J.C.H, Schlichting, I, Chapman, H.N, Fromme, P, Neutze, R. | | Deposit date: | 2013-10-09 | | Release date: | 2013-12-25 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Structure of a photosynthetic reaction centre determined by serial femtosecond crystallography.
Nat Commun, 4, 2013
|
|
8VLL
 
 | | Crystal structure of the yeast cytosine deaminase (yCD) M100W mutant | | Descriptor: | 1,2-ETHANEDIOL, Cytosine deaminase, PHOSPHATE ION, ... | | Authors: | Picard, M.-E, Grenier, J, Despres, P.C, Dube, A.K, Landry, C.R, Shi, R. | | Deposit date: | 2024-01-11 | | Release date: | 2024-08-21 | | Last modified: | 2024-09-18 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Compensatory mutations potentiate constructive neutral evolution by gene duplication.
Science, 385, 2024
|
|
8VLJ
 
 | | Crystal structure of the cacodylate-bound yeast cytosine deaminase (closed form) | | Descriptor: | 1,2-ETHANEDIOL, CACODYLATE ION, Cytosine deaminase, ... | | Authors: | Picard, M.-E, Grenier, J, Despres, P.C, Dube, A.K, Landry, C.R, Shi, R. | | Deposit date: | 2024-01-11 | | Release date: | 2024-08-21 | | Last modified: | 2024-09-04 | | Method: | X-RAY DIFFRACTION (1.39 Å) | | Cite: | Compensatory mutations potentiate constructive neutral evolution by gene duplication.
Science, 385, 2024
|
|
8VLK
 
 | | Crystal structure of the yeast cytosine deaminase containing both open and closed active sites | | Descriptor: | 1,2-ETHANEDIOL, Cytosine deaminase, SULFATE ION, ... | | Authors: | Picard, M.-E, Grenier, J, Despres, P.C, Dube, A.K, Landry, C.R, Shi, R. | | Deposit date: | 2024-01-11 | | Release date: | 2024-08-21 | | Last modified: | 2024-09-04 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Compensatory mutations potentiate constructive neutral evolution by gene duplication.
Science, 385, 2024
|
|
4ET9
 
 | | Hen egg-white lysozyme solved from 5 fs free-electron laser pulse data | | Descriptor: | CHLORIDE ION, Lysozyme C, SODIUM ION | | Authors: | Boutet, S, Lomb, L, Williams, G, Barends, T, Aquila, A, Doak, R.B, Weierstall, U, DePonte, D, Steinbrener, J, Shoeman, R, Messerschmidt, M, Barty, A, White, T, Kassemeyer, S, Kirian, R, Seibert, M, Montanez, P, Kenney, C, Herbst, R, Hart, P, Pines, J, Haller, G, Gruner, S, Philllip, H, Tate, M, Hromalik, M, Koerner, L, van Bakel, N, Morse, J, Ghonsalves, W, Arnlund, D, Bogan, M, Calemann, C, Fromme, R, Hampton, C, Hunter, M, Johansson, L, Katona, G, Kupitz, C, Liang, M, Martin, A, Nass, K, Redecke, L, Stellato, F, Timneanu, N, Wang, D, Zatsepin, N, Schafer, D, Defever, K, Neutze, R, Fromme, P, Spence, J, Chapman, H, Schlichting, I. | | Deposit date: | 2012-04-24 | | Release date: | 2012-06-13 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | High-resolution protein structure determination by serial femtosecond crystallography.
Science, 337, 2012
|
|
