2W8T
 
 | | SPT with PLP, N100C | | Descriptor: | PYRIDOXAL-5'-PHOSPHATE, SERINE PALMITOYLTRANSFERASE | | Authors: | Raman, M.C.C, Johnson, K.A, Campopiano, D.J, Naismith, J.H. | | Deposit date: | 2009-01-19 | | Release date: | 2009-01-27 | | Last modified: | 2025-04-09 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | The External-Aldimine Form of Serine Palmitoyltranserase; Structural, Kinetic and Spectroscopic Analysis of the Wild-Type Enzyme and Hsan1 Mutant Mimics.
J.Biol.Chem., 284, 2009
|
|
2W8U
 
 | | SPT with PLP, N100Y | | Descriptor: | PYRIDOXAL-5'-PHOSPHATE, SERINE PALMITOYLTRANSFERASE | | Authors: | Raman, M.C.C, Johnson, K.A, Campopiano, D.J, Naismith, J.H. | | Deposit date: | 2009-01-19 | | Release date: | 2009-01-27 | | Last modified: | 2025-04-09 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The External-Aldimine Form of Serine Palmitoyltranserase; Structural, Kinetic and Spectroscopic Analysis of the Wild-Type Enzyme and Hsan1 Mutant Mimics.
J.Biol.Chem., 284, 2009
|
|
2W8V
 
 | | SPT with PLP, N100W | | Descriptor: | PYRIDOXAL-5'-PHOSPHATE, SERINE PALMITOYLTRANSFERASE | | Authors: | Raman, M.C.C, Johnson, K.A, Campopiano, D.J, Naismith, J.H. | | Deposit date: | 2009-01-19 | | Release date: | 2009-01-27 | | Last modified: | 2025-04-09 | | Method: | X-RAY DIFFRACTION (1.43 Å) | | Cite: | The External-Aldimine Form of Serine Palmitoyltranserase; Structural, Kinetic and Spectroscopic Analysis of the Wild-Type Enzyme and Hsan1 Mutant Mimics.
J.Biol.Chem., 284, 2009
|
|
2X0D
 
 | | APO structure of WsaF | | Descriptor: | GLYCEROL, WSAF | | Authors: | Steiner, K, Hagelueken, G, Naismith, J.H. | | Deposit date: | 2009-12-08 | | Release date: | 2010-02-02 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Structural Basis of Substrate Binding in Wsaf, a Rhamnosyltransferase from Geobacillus Stearothermophilus.
J.Mol.Biol., 397, 2010
|
|
2X6T
 
 | | AGME bound to ADP-B-mannose | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADP-L-GLYCERO-D-MANNO-HEPTOSE-6-EPIMERASE, CHLORIDE ION, ... | | Authors: | Kowatz, T, Morrison, J.P, Tanner, M.E, Naismith, J.H. | | Deposit date: | 2010-02-21 | | Release date: | 2010-03-02 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.36 Å) | | Cite: | The Crystal Structure of the Y140F Mutant of Adp-L-Glycero-D-Manno-Heptose 6-Epimerase Bound to Adp-Beta-D-Mannose Suggests a One Base Mechanism.
Protein Sci., 19, 2010
|
|
2X48
 
 | | ORF 55 from Sulfolobus islandicus rudivirus 1 | | Descriptor: | CAG38821, PHOSPHATE ION | | Authors: | Oke, M, Carter, L, Johnson, K.A, Liu, H, Mcmahon, S, Naismith, J.H, White, M.F. | | Deposit date: | 2010-01-28 | | Release date: | 2010-07-21 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The Scottish Structural Proteomics Facility: Targets, Methods and Outputs.
J.Struct.Funct.Genomics, 11, 2010
|
|
2X0O
 
 | | Apo structure of the Alcaligin biosynthesis protein C (AlcC) from Bordetella bronchiseptica | | Descriptor: | ALCALIGIN BIOSYNTHESIS PROTEIN, SULFATE ION | | Authors: | Johnson, K.A, Schmelz, S, Kadi, N, Mcmahon, S.A, Oke, M, Liu, H, Carter, L.G, White, M.F, Challis, G.L, Naismith, J.H. | | Deposit date: | 2009-12-16 | | Release date: | 2010-07-28 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The Scottish Structural Proteomics Facility: Targets, Methods and Outputs.
J.Struct.Funct.Genomics, 11, 2010
|
|
2WEU
 
 | |
8QXC
 
 | | Crystal structure of antibody Fab MIL-3 with PenG-Lys | | Descriptor: | (2R,4S)-2-[(1R)-2-[[(5S)-5-acetamido-6-oxidanyl-6-oxidanylidene-hexyl]amino]-2-oxidanylidene-1-(2-phenylethanoylamino)ethyl]-5,5-dimethyl-1,3-thiazolidine-4-carboxylic acid, Heavy chain of FAB MIL-3, Light chain of FAB MIL-3 | | Authors: | Moynie, L, Naismith, J.H. | | Deposit date: | 2023-10-24 | | Release date: | 2024-07-31 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Covalent penicillin-protein conjugates elicit anti-drug antibodies that are clonally and functionally restricted.
Nat Commun, 15, 2024
|
|
6T38
 
 | | Pseudomonas aeruginosa RmlA in complex with allosteric inhibitor | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, BROMIDE ION, CHLORIDE ION, ... | | Authors: | Alphey, M.S, Xiao, G, Westwood, J.N. | | Deposit date: | 2019-10-10 | | Release date: | 2020-08-19 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Next generation Glucose-1-phosphate thymidylyltransferase (RmlA) inhibitors: An extended SAR study to direct future design.
Bioorg.Med.Chem., 50, 2021
|
|
6T37
 
 | | Pseudomonas aeruginosa RmlA in complex with allosteric inhibitor | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CHLORIDE ION, Glucose-1-phosphate thymidylyltransferase, ... | | Authors: | Alphey, M.S, Xiao, G, Westwood, J.N. | | Deposit date: | 2019-10-10 | | Release date: | 2020-08-19 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.079 Å) | | Cite: | Next generation Glucose-1-phosphate thymidylyltransferase (RmlA) inhibitors: An extended SAR study to direct future design.
Bioorg.Med.Chem., 50, 2021
|
|
6TQG
 
 | | Pseudomonas aeruginosa RmlA in complex with allosteric inhibitor | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CHLORIDE ION, Glucose-1-phosphate thymidylyltransferase, ... | | Authors: | Alphey, M.S, Xiao, G, Westwood, J.N. | | Deposit date: | 2019-12-16 | | Release date: | 2020-08-19 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Next generation Glucose-1-phosphate thymidylyltransferase (RmlA) inhibitors: An extended SAR study to direct future design.
Bioorg.Med.Chem., 50, 2021
|
|
4ETR
 
 | | X-ray structure of PA2169 from Pseudomonas aeruginosa | | Descriptor: | Putative uncharacterized protein | | Authors: | Schnell, R, Sandalova, T, Lindqvist, Y, Schneider, G. | | Deposit date: | 2012-04-24 | | Release date: | 2013-01-09 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | The AEROPATH project targeting Pseudomonas aeruginosa: crystallographic studies for assessment of potential targets in early-stage drug discovery.
Acta Crystallogr.,Sect.F, 69, 2013
|
|
4ES6
 
 | |
4EXB
 
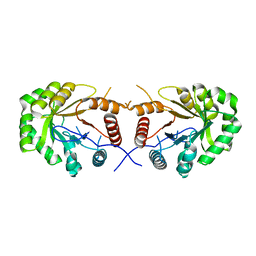 | |
4EXA
 
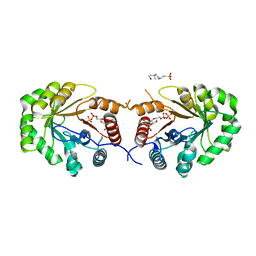 | | Crystal structure of the PA4992, the putative aldo-keto reductase from Pseudomona aeruginosa | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Putative uncharacterized protein | | Authors: | Sandalova, T, Schnell, R, Schneider, G. | | Deposit date: | 2012-04-30 | | Release date: | 2013-01-09 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The AEROPATH project targeting Pseudomonas aeruginosa: crystallographic studies for assessment of potential targets in early-stage drug discovery.
Acta Crystallogr.,Sect.F, 69, 2013
|
|
3D4K
 
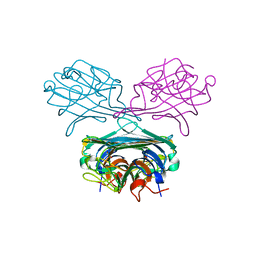 | | Concanavalin A Complexed to a Synthetic Analog of the Trimannoside | | Descriptor: | CALCIUM ION, Concanavalin-A, GLYCEROL, ... | | Authors: | Kadirvelraj, R, Foley, B.L, Dyekjaer, J.D, Woods, R.J. | | Deposit date: | 2008-05-14 | | Release date: | 2009-03-03 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Involvement of water in carbohydrate-protein binding: concanavalin A revisited.
J.Am.Chem.Soc., 130, 2008
|
|
6ENE
 
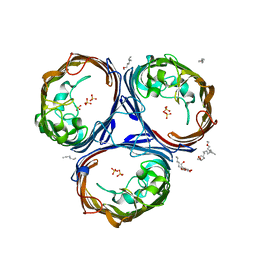 | | OmpF orthologue from Enterobacter cloacae (OmpE35) | | Descriptor: | (HYDROXYETHYLOXY)TRI(ETHYLOXY)OCTANE, Outer membrane protein (Porin), SULFATE ION, ... | | Authors: | van den Berg, B, Abellon-Ruiz, J, Basle, A. | | Deposit date: | 2017-10-04 | | Release date: | 2018-10-31 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Getting Drugs into Gram-Negative Bacteria: Rational Rules for Permeation through General Porins.
Acs Infect Dis., 4, 2018
|
|
8B72
 
 | |
6ZFO
 
 | | Association of two complexes of largely structurally disordered Spike ectodomain with bound EY6A Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, EY6A heavy chain, EY6A light chain, ... | | Authors: | Duyvesteyn, H.M.E, Zhou, D, Zhao, Y, Fry, E.E, Ren, J, Stuart, D.I. | | Deposit date: | 2020-06-17 | | Release date: | 2020-07-08 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (4.4 Å) | | Cite: | Structural basis for the neutralization of SARS-CoV-2 by an antibody from a convalescent patient.
Nat.Struct.Mol.Biol., 27, 2020
|
|
6ZDH
 
 | | SARS-CoV-2 Spike glycoprotein in complex with a neutralizing antibody EY6A Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, EY6A heavy chain, ... | | Authors: | Duyvesteyn, H.M.E, Zhou, D, Zhao, Y, Fry, E.E, Ren, J, Stuart, D.I. | | Deposit date: | 2020-06-14 | | Release date: | 2020-07-01 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Structural basis for the neutralization of SARS-CoV-2 by an antibody from a convalescent patient.
Nat.Struct.Mol.Biol., 27, 2020
|
|
6QD6
 
 | | Molecular scaffolds expand the nanobody toolkit for cryo-EM applications: crystal structure of Mb-cHopQ-Nb207 | | Descriptor: | CHLORIDE ION, Mb-cHopQ-Nb207,Outer membrane protein,Mb-cHopQ-Nb207,Outer membrane protein,Mb-cHopQ-Nb207 | | Authors: | Uchanski, T, Masiulis, S, Fischer, B, Kalichuk, V, Wohlkonig, A, Zogg, T, Remaut, H, Vranken, W, Aricescu, A.R, Pardon, E, Steyaert, J. | | Deposit date: | 2018-12-31 | | Release date: | 2019-12-18 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.84 Å) | | Cite: | Megabodies expand the nanobody toolkit for protein structure determination by single-particle cryo-EM
Nat.Methods, 18, 2021
|
|
6XVI
 
 | |
6XUX
 
 | | Crystal structure of Megabody Mb-Nb207-cYgjK_NO | | Descriptor: | CALCIUM ION, Nanobody,Glucosidase YgjK,Glucosidase YgjK,Nanobody | | Authors: | Steyaert, J, Uchanski, T, Fischer, B. | | Deposit date: | 2020-01-21 | | Release date: | 2021-01-13 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.90000641 Å) | | Cite: | Megabodies expand the nanobody toolkit for protein structure determination by single-particle cryo-EM.
Nat.Methods, 18, 2021
|
|
6XV8
 
 | |
