4BQG
 
 | | structure of HSP90 with an inhibitor bound | | Descriptor: | 5-(3,4-dichloro-phenoxy)-benzene-1,3-diol, HSP90AA1 PROTEIN | | Authors: | Casale, E, Brasca, M.G, Mantegani, S, Amboldi, N, Bindi, S, Caronni, D, Ceccarelli, W, Colombo, N, DePonti, A, Donati, D, Ermoli, A, Fachin, G, Felder, E.R, Ferguson, R.D, Fiorelli, C, Guanci, M, Isacchi, A, Pesenti, E, Polucci, P, Riceputi, L, Sola, F, Visco, C, Zuccotto, F, Fogliatto, G. | | Deposit date: | 2013-05-30 | | Release date: | 2013-10-23 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Discovery of Nms-E973 as Novel, Selective and Potent Inhibitor of Heat Shock Protein 90 (Hsp90).
Bioorg.Med.Chem., 21, 2013
|
|
7KP8
 
 | | asymmetric mTNF-alpha hTNFR1 complex | | Descriptor: | 1-{[2-(difluoromethoxy)phenyl]methyl}-2-methyl-6-[6-(piperazin-1-yl)pyridin-3-yl]-1H-benzimidazole, Tumor necrosis factor, Tumor necrosis factor receptor superfamily member 1A | | Authors: | Arakaki, T.L, Fox III, D, Edwards, T.E, Foley, A, Ceska, T. | | Deposit date: | 2020-11-10 | | Release date: | 2021-01-13 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | Structural insights into the disruption of TNF-TNFR1 signalling by small molecules stabilising a distorted TNF.
Nat Commun, 12, 2021
|
|
3FL1
 
 | | X-ray structure of the non covalent swapped form of the A19P/Q28L/K31C/S32C mutant of bovine pancreatic ribonuclease in complex with 2'-DEOXYCYTIDINE-2'-DEOXYGUANOSINE-3',5'-MONOPHOSPHATE | | Descriptor: | 2'-DEOXYCYTIDINE-2'-DEOXYGUANOSINE-3',5'-MONOPHOSPHATE, Ribonuclease pancreatic, SULFATE ION, ... | | Authors: | Merlino, A, Russo Krauss, I, Perillo, M, Mattia, C.A, Ercole, C, Picone, D, Vergara, A, Sica, F. | | Deposit date: | 2008-12-18 | | Release date: | 2009-03-24 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Toward an antitumor form of bovine pancreatic ribonuclease: The crystal structure of three noncovalent dimeric mutants
Biopolymers, 91, 2009
|
|
7KKL
 
 | | SARS-CoV-2 Spike in complex with neutralizing nanobody mNb6 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, ... | | Authors: | Schoof, M.S, Faust, B.F, Saunders, R.A, Sangwan, S, Rezelj, V, Hoppe, N, Boone, M, Billesboelle, C.B, Puchades, C, Azumaya, C.M, Kratochvil, H.T, Zimanyi, M, Desphande, I, Liang, J, Dickinson, S, Nguyen, H.C, Chio, C.M, Merz, G.E, Thompson, M.C, Diwanji, D, Schaefer, K, Anand, A.A, Dobzinski, N, Zha, B.S, Simoneau, C.R, Leon, K, White, K.M, Chio, U.S, Gupta, M, Jin, M, Li, F, Liu, Y, Zhang, K, Bulkley, D, Sun, M, Smith, A.M, Rizo, A.N, Moss, F, Brilot, A.F, Pourmal, S, Trenker, R, Pospiech, T, Gupta, S, Barsi-Rhyne, B, Belyy, V, Barile-Hill, A.W, Nock, S, Liu, Y, Krogan, N.J, Ralston, C.Y, Swaney, D.L, Garcia-Sastre, A, Ott, M, Vignuzzi, M, Walter, P, Manglik, A, QCRG Structural Biology Consortium | | Deposit date: | 2020-10-27 | | Release date: | 2020-11-11 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (2.85 Å) | | Cite: | An ultrapotent synthetic nanobody neutralizes SARS-CoV-2 by stabilizing inactive Spike.
Science, 370, 2020
|
|
4BZN
 
 | | Crystal structure of PIM1 in complex with a Pyrrolo(1,2-a)Pyrazinone inhibitor | | Descriptor: | N-(2,2-dimethylpropyl)-2-[1-oxo-7-(thiophen-3-yl)-1,2,3,4-tetrahydropyrrolo[1,2-a]pyrazin-4-yl]acetamide, SERINE/THREONINE-PROTEIN KINASE PIM-1 | | Authors: | Casale, E, Casuscelli, F, Ardini, E, Avanzi, N, Cervi, G, D'Anello, M, Donati, D, Faiardi, D, Ferguson, R.D, Fogliatto, G, Galvani, A, Marsiglio, A, Mirizzi, D.G, Montemartini, M, Orrenius, C, Papeo, G, Piutti, C, Salom, B, Felder, E.R. | | Deposit date: | 2013-07-29 | | Release date: | 2013-10-30 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Discovery and Optimization of Pyrrolo[1,2-A]Pyrazinones Leads to Novel and Selective Inhibitors of Pim Kinases.
Bioorg.Med.Chem., 21, 2013
|
|
1ZK4
 
 | | Structure of R-specific alcohol dehydrogenase (wildtype) from Lactobacillus brevis in complex with acetophenone and NADP | | Descriptor: | 1-PHENYLETHANONE, MAGNESIUM ION, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Schlieben, N.H, Niefind, K, Muller, J, Riebel, B, Hummel, W, Schomburg, D. | | Deposit date: | 2005-05-02 | | Release date: | 2005-06-21 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Atomic Resolution Structures of R-specific Alcohol Dehydrogenase from Lactobacillus brevis Provide the Structural Bases of its Substrate and Cosubstrate Specificity
J.Mol.Biol., 349, 2005
|
|
1XYM
 
 | | THE ROLE OF THE DIVALENT METAL ION IN SUGAR BINDING, RING OPENING, AND ISOMERIZATION BY D-XYLOSE ISOMERASE: REPLACEMENT OF A CATALYTIC METAL BY AN AMINO-ACID | | Descriptor: | D-glucose, HYDROXIDE ION, MAGNESIUM ION, ... | | Authors: | Allen, K.N, Lavie, A, Petsko, G.A, Ringe, D. | | Deposit date: | 1993-12-07 | | Release date: | 1994-05-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Role of the divalent metal ion in sugar binding, ring opening, and isomerization by D-xylose isomerase: replacement of a catalytic metal by an amino acid.
Biochemistry, 33, 1994
|
|
4BYG
 
 | | ATPase crystal structure | | Descriptor: | COPPER EFFLUX ATPASE, MAGNESIUM ION, POLYETHYLENE GLYCOL (N=34), ... | | Authors: | Mattle, D, Drachmann, N.D, Liu, X.Y, Pedersen, B.P, Morth, J.P, Wang, J, Gourdon, P, Nissen, P. | | Deposit date: | 2013-07-19 | | Release date: | 2014-08-06 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Dephosphorylation of Pib-Type Cu(I)-Atpases as Studied by Metallofluoride Complexes
To be Published
|
|
9AT8
 
 | | Fab 77-stabilized MeV F ectodomain fragment | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Fusion glycoprotein F0, mAB 77 heavy chain, ... | | Authors: | Zyla, D, Saphire, E.O. | | Deposit date: | 2024-02-26 | | Release date: | 2024-07-03 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | A neutralizing antibody prevents postfusion transition of measles virus fusion protein.
Science, 384, 2024
|
|
3FPV
 
 | | Crystal Structure of HbpS | | Descriptor: | Extracellular haem-binding protein, FE (III) ION | | Authors: | Ortiz de Orue Lucana, D, Bogel, G, Zou, P, Groves, M.R. | | Deposit date: | 2009-01-06 | | Release date: | 2009-01-20 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The oligomeric assembly of the novel haem-degrading protein HbpS is essential for interaction with its cognate two-component sensor kinase
J.Mol.Biol., 386, 2009
|
|
1FCZ
 
 | | ISOTYPE SELECTIVITY OF THE HUMAN RETINOIC ACID NUCLEAR RECEPTOR HRAR: THE COMPLEX WITH THE PANAGONIST RETINOID BMS181156 | | Descriptor: | 4-[3-OXO-3-(5,5,8,8-TETRAMETHYL-5,6,7,8-TETRAHYDRO-NAPHTHALEN-2-YL)-PROPENYL]-BENZOIC ACID, DODECYL-ALPHA-D-MALTOSIDE, RETINOIC ACID RECEPTOR GAMMA-1 | | Authors: | Klaholz, B.P, Mitschler, A, Moras, D, Structural Proteomics in Europe (SPINE) | | Deposit date: | 2000-07-19 | | Release date: | 2000-09-11 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.38 Å) | | Cite: | Structural basis for isotype selectivity of the human retinoic acid nuclear receptor.
J.Mol.Biol., 302, 2000
|
|
7DFV
 
 | | Cryo-EM structure of plant NLR RPP1 tetramer core part | | Descriptor: | NAD+ hydrolase (NADase) | | Authors: | Ma, S.C, Lapin, D, Liu, L, Sun, Y, Song, W, Zhang, X.X, Logemann, E, Yu, D.L, Wang, J, Jirschitzka, J, Han, Z.F, SchulzeLefert, P, Parker, J.E, Chai, J.J. | | Deposit date: | 2020-11-10 | | Release date: | 2020-12-16 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (2.99 Å) | | Cite: | Direct pathogen-induced assembly of an NLR immune receptor complex to form a holoenzyme.
Science, 370, 2020
|
|
1LEN
 
 | | REFINEMENT OF TWO CRYSTAL FORMS OF LENTIL LECTIN AT 1.8 ANGSTROMS RESOLUTION | | Descriptor: | CALCIUM ION, LECTIN, MANGANESE (II) ION, ... | | Authors: | Van Overberge, D, Loris, R, Wyns, L. | | Deposit date: | 1993-11-17 | | Release date: | 1994-01-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural analysis of two crystal forms of lentil lectin at 1.8 A resolution.
Proteins, 20, 1994
|
|
5NJM
 
 | | Lysozyme room-temperature structure determined by serial millisecond crystallography | | Descriptor: | Lysozyme C | | Authors: | Weinert, T, Vera, L, Marsh, M, James, D, Gashi, D, Nogly, P, Jaeger, K, Standfuss, J. | | Deposit date: | 2017-03-29 | | Release date: | 2017-09-27 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Serial millisecond crystallography for routine room-temperature structure determination at synchrotrons.
Nat Commun, 8, 2017
|
|
6DM9
 
 | | DHD15_extended | | Descriptor: | DHD15_extended_A, DHD15_extended_B, SULFATE ION | | Authors: | Bick, M.J, Chen, Z, Baker, D. | | Deposit date: | 2018-06-04 | | Release date: | 2018-12-19 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Programmable design of orthogonal protein heterodimers.
Nature, 565, 2019
|
|
6DKM
 
 | | DHD131 | | Descriptor: | DHD131_A, DHD131_B | | Authors: | Bick, M.J, Chen, Z, Baker, D. | | Deposit date: | 2018-05-29 | | Release date: | 2018-12-19 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | Programmable design of orthogonal protein heterodimers.
Nature, 565, 2019
|
|
1XYB
 
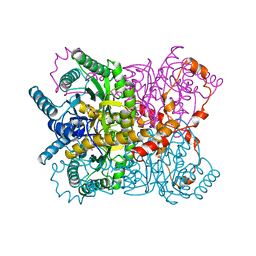 | | X-RAY CRYSTALLOGRAPHIC STRUCTURES OF D-XYLOSE ISOMERASE-SUBSTRATE COMPLEXES POSITION THE SUBSTRATE AND PROVIDE EVIDENCE FOR METAL MOVEMENT DURING CATALYSIS | | Descriptor: | D-glucose, MAGNESIUM ION, XYLOSE ISOMERASE | | Authors: | Lavie, A, Allen, K.N, Petsko, G.A, Ringe, D. | | Deposit date: | 1994-01-03 | | Release date: | 1994-05-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | X-ray crystallographic structures of D-xylose isomerase-substrate complexes position the substrate and provide evidence for metal movement during catalysis.
Biochemistry, 33, 1994
|
|
6S2Y
 
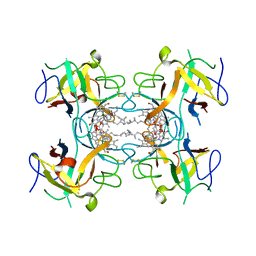 | | Water-soluble Chlorophyll Protein (WSCP) from Lepidium virginicum with Chlorophyll-b | | Descriptor: | CHLOROPHYLL B, Water-soluble chlorophyll protein | | Authors: | Agostini, A, Meneghin, E, Gewehr, L, Pedron, D, Palm, D.M, Carbonera, D, Paulsen, H, Jaenicke, E, Collini, E. | | Deposit date: | 2019-06-23 | | Release date: | 2019-12-18 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | How water-mediated hydrogen bonds affect chlorophyll a/b selectivity in Water-Soluble Chlorophyll Protein.
Sci Rep, 9, 2019
|
|
7D2Z
 
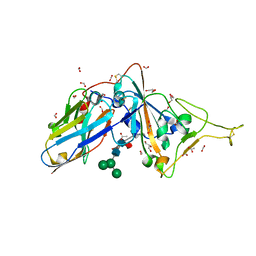 | | Structure of sybody SR31 in complex with the SARS-CoV-2 S Receptor Binding domain (RBD) | | Descriptor: | ACETATE ION, FORMIC ACID, GLYCEROL, ... | | Authors: | Li, T, Cai, H, Yao, H, Qin, W, Li, D. | | Deposit date: | 2020-09-17 | | Release date: | 2021-02-17 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | A high-affinity RBD-targeting nanobody improves fusion partner's potency against SARS-CoV-2.
Plos Pathog., 17, 2021
|
|
7D30
 
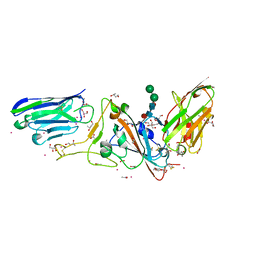 | | Structure of sybody MR17-SR31 fusion in complex with the SARS-CoV-2 S Receptor Binding domain (RBD) | | Descriptor: | 1,3-BUTANEDIOL, ACETATE ION, CADMIUM ION, ... | | Authors: | Li, T, Yao, H, Cai, H, Qin, W, Li, D. | | Deposit date: | 2020-09-17 | | Release date: | 2021-02-17 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | A high-affinity RBD-targeting nanobody improves fusion partner's potency against SARS-CoV-2.
Plos Pathog., 17, 2021
|
|
6RJ1
 
 | |
4BVC
 
 | | Identification of small molecule inhibitors selective for apo(a) kringles KIV-7, KIV-10 and KV. | | Descriptor: | 3-(4-PIPERIDYL)-N-[2-(TRIFLUOROMETHOXY)PHENYL]SULFONYL-PROPANAMIDE, APOLIPOPROTEIN(A), CHLORIDE ION, ... | | Authors: | Sandmark, J, Althage, M, Andersson, G.M.K, Antonsson, T, Blaho, S, Bodin, C, Bostrom, J, Chen, Y, Dahlen, A, Eriksson, P.O, Evertsson, E, Fex, T, Fjellstrom, O, Gustafsson, D, Hallberg, C, Hicks, R, Jarkvist, E, Johansson, C, Kalies, I, Kang, D, Svalstedt Karlsson, B, Kartberg, F, Legnehed, A, Lindqvist, A.M, Martinsson, S.A, Moberg, A, Petersson, A.U, Ridderstrom, M, Thelin, A, Tigerstrom, A, Vinblad, J, Xu, B, Knecht, W. | | Deposit date: | 2013-06-25 | | Release date: | 2014-07-09 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Small Molecules Used to Decipher the Pathophysiological Roles of the Kringle Domains Kiv-7, - 10 and Kv of Apolipoprotein(A)
To be Published
|
|
3ZO8
 
 | | Wild-type chorismate mutase of Bacillus subtilis at 1.6 A resolution | | Descriptor: | CHORISMATE MUTASE AROH | | Authors: | Burschowsky, D, vanEerde, A, Okvist, M, Kienhofer, A, Kast, P, Hilvert, D, Krengel, U. | | Deposit date: | 2013-02-20 | | Release date: | 2014-04-16 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Electrostatic Transition State Stabilization Rather Than Reactant Destabilization Provides the Chemical Basis for Efficient Chorismate Mutase Catalysis.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
8AGH
 
 | | BK Polyomavirus VP1 mutant E73A | | Descriptor: | CHLORIDE ION, GLYCEROL, Major capsid protein VP1 | | Authors: | Sorin, M.N, Di Maio, A, Silva, L.M, Ebert, D, Delannoy, C, Nguyen, N.-K, Guerardel, Y, Chai, W, Halary, F, Renaudin-Autain, K, Liu, Y, Bressollette-Bodin, C, Stehle, T, McIlroy, D. | | Deposit date: | 2022-07-20 | | Release date: | 2023-02-22 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.887 Å) | | Cite: | Structural and functional analysis of natural capsid variants suggests sialic acid-independent entry of BK polyomavirus.
Cell Rep, 42, 2023
|
|
8AGO
 
 | | BK Polyomavirus VP1 mutant E73Q | | Descriptor: | CHLORIDE ION, GLYCEROL, Major capsid protein VP1 | | Authors: | Sorin, M.N, Di Maio, A, Silva, L.M, Ebert, D, Delannoy, C, Nguyen, N.-K, Guerardel, Y, Chai, W, Halary, F, Renaudin-Autain, K, Liu, Y, Bressollette-Bodin, C, Stehle, T, McIlroy, D. | | Deposit date: | 2022-07-20 | | Release date: | 2023-02-22 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.853 Å) | | Cite: | Structural and functional analysis of natural capsid variants suggests sialic acid-independent entry of BK polyomavirus.
Cell Rep, 42, 2023
|
|
