7LH8
 
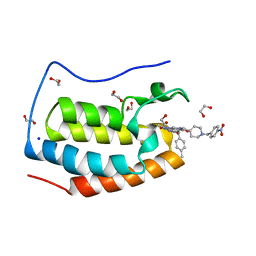 | | CRYSTAL STRUCTURE OF THE FIRST BROMODOMAIN OF HUMAN BRD4 IN COMPLEX WITH INHIBITOR JJ-II-131 | | Descriptor: | (5R)-1-ethyl-5-(4-methylphenyl)-7-({1-[(4-nitrophenyl)methyl]piperidin-4-yl}methyl)-5,7,8,9-tetrahydro-1H-pyrrolo[3',4':5,6]pyrido[2,3-d]pyrimidine-2,4,6(3H)-trione, 1,2-ETHANEDIOL, Bromodomain-containing protein 4, ... | | Authors: | Schonbrunn, E, Chan, A. | | Deposit date: | 2021-01-21 | | Release date: | 2022-07-06 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Dihydropyridine Lactam Analogs Targeting BET Bromodomains.
Chemmedchem, 17, 2022
|
|
7DLQ
 
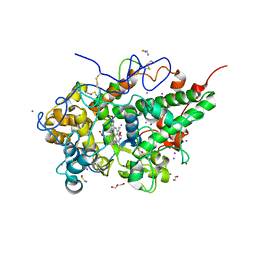 | | CRYSTAL STRUCTURE OF THE COMPLEX OF LACTOPEROXIDASE WITH HYDROGEN PEROXIDE AT 1.77A RESOLUTION | | Descriptor: | 1,2-ETHANEDIOL, 1-(OXIDOSULFANYL)METHANAMINE, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Singh, P.K, Sharma, P, Bhushan, A, Sharma, S, Singh, T.P. | | Deposit date: | 2020-11-29 | | Release date: | 2020-12-16 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.773 Å) | | Cite: | Structure of a ternary complex of lactoperoxidase with iodide and hydrogen peroxide at 1.77 angstrom resolution.
J.Inorg.Biochem., 220, 2021
|
|
2CEN
 
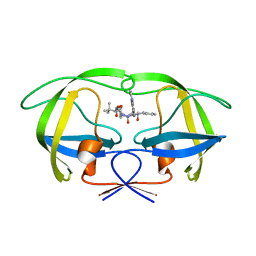 | | P1' Extended HIV-1 Protease Inhibitors Encompassing a Tertiary Alcohol in the Transition-State Mimicking Scaffold | | Descriptor: | POL PROTEIN, {(1S)-1-[N'-[(2S)-2-HYDROXY-2-((1S,2R)-2-HYDROXY-INDAN-1-YLCARBAMOYL)-3-PHENYL-PROPYL]-N'-[4-(PYRIDINE-2-YL)-BENZYL]-HYDRAZINOCARBONYL]-2,2-DIMETHYL-PROPYL}-CARBAMIC ACID METHYL ESTER | | Authors: | Ginman, N, Ekegren, J.K, Johansson, A, Wallberg, H, Larhed, M, Samuelsson, B, Hallberg, A, Unge, T. | | Deposit date: | 2006-02-08 | | Release date: | 2007-02-13 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Microwave-Accelerated Synthesis of P1'-Extended HIV-1 Protease Inhibitors Encompassing a Tertiary Alcohol in the Transition-State Mimicking Scaffold.
J.Med.Chem., 49, 2006
|
|
3FXT
 
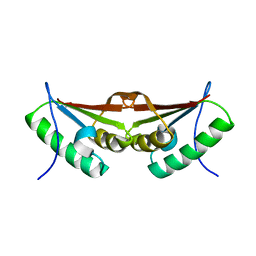 | | Crystal structure of the N-terminal domain of human NUDT6 | | Descriptor: | GLYCEROL, Nucleoside diphosphate-linked moiety X motif 6 | | Authors: | Tresaugues, L, Welin, M, Arrowsmith, C.H, Berglund, H, Bountra, C, Collins, R, Dahlgren, L.G, Edwards, A.M, Flodin, S, Flores, A, Graslund, S, Hammarstrom, M, Johansson, A, Johansson, I, Karlberg, T, Kotenyova, T, Lehtio, L, Moche, M, Nilsson, M.E, Nyman, T, Persson, C, Sagemark, J, Schueler, H, Siponen, M.I, Thorsell, A.G, Van Den Berg, S, Weigelt, J, Wikstrom, M, Nordlund, P, Structural Genomics Consortium (SGC) | | Deposit date: | 2009-01-21 | | Release date: | 2009-03-03 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of the N-terminal domain of human NUDT6
To be Published
|
|
3G1U
 
 | | Crystal structure of Leishmania major S-adenosylhomocysteine hydrolase | | Descriptor: | ADENOSINE, Adenosylhomocysteinase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Siponen, M.I, Welin, M, Arrowsmith, C.H, Berglund, H, Bountra, C, Collins, R, Dahlgren, L.G, Edwards, A.M, Flodin, S, Flores, A, Graslund, S, Hammarstrom, M, Johansson, A, Johansson, I, Karlberg, T, Kotenyova, T, Lehtio, L, Moche, M, Nilsson, M.E, Nordlund, P, Nyman, T, Persson, C, Sagemark, J, Schutz, P, Thorsell, A.G, Tresaugues, L, Van Den Berg, S, Weigelt, J, Wisniewska, M, Schueler, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2009-01-30 | | Release date: | 2009-02-10 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of Leishmania major S-adenosylhomocysteine hydrolase
To be Published
|
|
6GS1
 
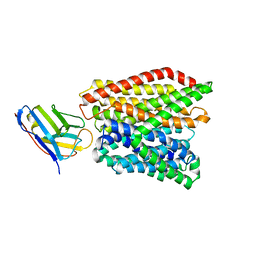 | | Crystal structure of peptide transporter DtpA-nanobody in MES buffer | | Descriptor: | Dipeptide and tripeptide permease A, Nanobody 00 | | Authors: | Ural-Blimke, Y, Flayhan, A, Loew, C, Quistgaard, E.M. | | Deposit date: | 2018-06-13 | | Release date: | 2019-01-30 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.29 Å) | | Cite: | Structure of Prototypic Peptide Transporter DtpA from E. coli in Complex with Valganciclovir Provides Insights into Drug Binding of Human PepT1.
J. Am. Chem. Soc., 141, 2019
|
|
6GS4
 
 | | Crystal structure of peptide transporter DtpA-nanobody in complex with valganciclovir | | Descriptor: | DODECYL-BETA-D-MALTOSIDE, Dipeptide and tripeptide permease A, [(2~{S})-2-[(2-azanyl-6-oxidanylidene-3~{H}-purin-9-yl)methoxy]-3-oxidanyl-propyl] (2~{S})-2-azanyl-3-methyl-butanoate, ... | | Authors: | Ural-Blimke, Y, Flayhan, A, Quistgaard, E.M, Loew, C. | | Deposit date: | 2018-06-13 | | Release date: | 2019-01-30 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.645 Å) | | Cite: | Structure of Prototypic Peptide Transporter DtpA from E. coli in Complex with Valganciclovir Provides Insights into Drug Binding of Human PepT1.
J. Am. Chem. Soc., 141, 2019
|
|
6GS7
 
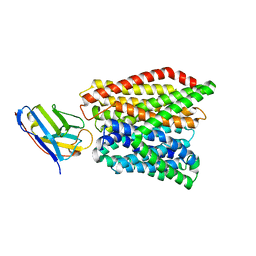 | | Crystal structure of peptide transporter DtpA-nanobody in glycine buffer | | Descriptor: | Dipeptide and tripeptide permease A, nanobody | | Authors: | Ural-Blimke, Y, Flayhan, A, Quistgaard, E.M, Loew, C. | | Deposit date: | 2018-06-13 | | Release date: | 2019-01-30 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structure of Prototypic Peptide Transporter DtpA from E. coli in Complex with Valganciclovir Provides Insights into Drug Binding of Human PepT1.
J. Am. Chem. Soc., 141, 2019
|
|
5J9G
 
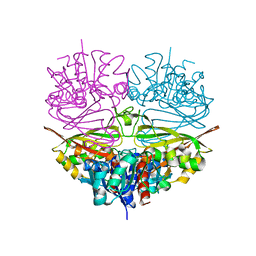 | |
2VXO
 
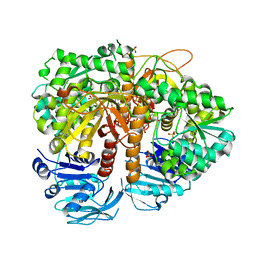 | | Human GMP synthetase in complex with XMP | | Descriptor: | GMP SYNTHASE [GLUTAMINE-HYDROLYZING], SULFATE ION, XANTHOSINE-5'-MONOPHOSPHATE | | Authors: | Welin, M, Lehtio, L, Andersson, J, Arrowsmith, C.H, Berglund, H, Collins, R, Dahlgren, L.G, Edwards, A.M, Flodin, S, Flores, A, Graslund, S, Hammarstrom, M, Johansson, A, Johansson, I, Karlberg, T, Kotenyova, T, Moche, M, Nilsson, M.E, Nyman, T, Olesen, K, Persson, C, Sagemark, J, Schueler, H, Thorsell, A.G, Tresaugues, L, Van Den Berg, S, Wisniewska, M, Wikstrom, M, Nordlund, P. | | Deposit date: | 2008-07-08 | | Release date: | 2008-08-12 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Substrate Specificity and Oligomerization of Human Gmp Synthetase
J.Mol.Biol., 425, 2013
|
|
2CEM
 
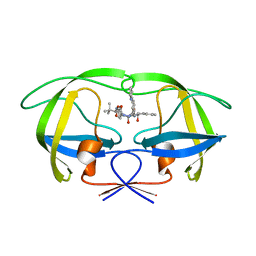 | | P1' Extended HIV-1 Protease Inhibitors Encompassing a Tertiary Alcohol in the Transition-State Mimicking Scaffold | | Descriptor: | POL PROTEIN, {(1S)-1-[N'-[(2S)-2-HYDROXY-2-((1S,2R)-2-HYDROXY-INDAN-1-YLCARBAMOYL)-3-PHENYL-PROPYL]-N'-[4-(PYRIDINE-2-YL)-BENZYL]-HYDRAZINOCARBONYL]-2,2-DIMETHYL-PROPYL}-CARBAMIC ACID METHYL ESTER | | Authors: | Ginman, N, Ekegren, J.K, Johansson, A, Wallberg, H, Larhed, M, Samuelsson, B, Hallberg, A, Unge, T. | | Deposit date: | 2006-02-08 | | Release date: | 2007-02-13 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Microwave-Accelerated Synthesis of P1'-Extended HIV-1 Protease Inhibitors Encompassing a Tertiary Alcohol in the Transition-State Mimicking Scaffold.
J.Med.Chem., 49, 2006
|
|
2CEJ
 
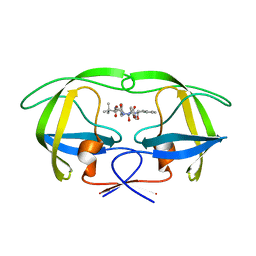 | | P1' Extended HIV-1 Protease Inhibitors Encompassing a Tertiary Alcohol in the Transition-State Mimicking Scaffold | | Descriptor: | 3-AMINO-3-BENZYL-[4.3.0]BICYCLO-1,6-DIAZANONAN-2-ONE, POL PROTEIN | | Authors: | Ginman, N, Ekegren, J.K, Johansson, A, Wallberg, H, Larhed, M, Samuelsson, B, Hallberg, A, Unge, T. | | Deposit date: | 2006-02-07 | | Release date: | 2007-02-13 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Microwave-Accelerated Synthesis of P1'-Extended HIV-1 Protease Inhibitors Encompassing a Tertiary Alcohol in the Transition-State Mimicking Scaffold
J.Med.Chem., 49, 2006
|
|
3F0W
 
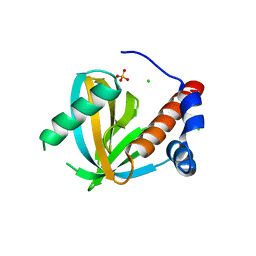 | | Human NUMB-like protein, phosphotyrosine interaction domain | | Descriptor: | CHLORIDE ION, Numb-like protein, SULFATE ION | | Authors: | Lehtio, L, Moche, M, Andersson, J, Arrowsmith, C.H, Berglund, H, Bountra, C, D Busam, R, Collins, R, Dahlgren, L.G, Edwards, A.M, Flodin, S, Flores, A, Graslund, S, Hammarstrom, M, Johansson, A, Johansson, I, Karlberg, T, Kotenyova, T, Nilsson, M.E, Nyman, T, Persson, C, Sagemark, J, Schueler, H, Thorsell, A.G, Tresaugues, L, Van Den Berg, S, Weigelt, J, Welin, M, Wikstrom, M, Wisniewska, M, Nordlund, P, Structural Genomics Consortium (SGC) | | Deposit date: | 2008-10-27 | | Release date: | 2008-11-04 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Human NUMB-like protein, phosphotyrosine interaction domain
To be Published
|
|
3GG6
 
 | | Crystal structure of the NUDIX domain of human NUDT18 | | Descriptor: | Nucleoside diphosphate-linked moiety X motif 18 | | Authors: | Tresaugues, L, Siponen, M.I, Lehtio, L, Arrowsmith, C.H, Berglund, H, Bountra, C, Collins, R, Dahlgren, L.G, Edwards, A.M, Flodin, S, Flores, A, Graslund, S, Hammarstrom, M, Johansson, A, Johansson, I, Karlberg, T, Kotenyova, T, Moche, M, Nilsson, M.E, Nyman, T, Persson, C, Sagemark, J, Schueler, H, Thorsell, A.G, Van Den Berg, S, Weigelt, J, Welin, M, Wisniewska, M, Nordlund, P, Structural Genomics Consortium (SGC) | | Deposit date: | 2009-02-27 | | Release date: | 2009-03-10 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of the NUDIX domain of human NUDT18
To be Published
|
|
6AHE
 
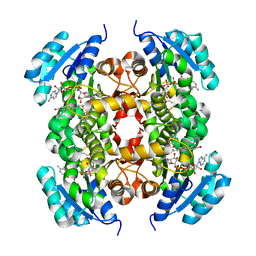 | | Crystal structure of enoyl-ACP reductase from Acinetobacter baumannii in complex with NAD and AFN-1252 | | Descriptor: | Enoyl-[acyl-carrier-protein] reductase [NADH], N-methyl-N-[(3-methyl-1-benzofuran-2-yl)methyl]-3-(7-oxo-5,6,7,8-tetrahydro-1,8-naphthyridin-3-yl)propanamide, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Rani, S.T, Nataraj, V, Laxminarasimhan, A, Thomas, A, Krishnamurthy, N. | | Deposit date: | 2018-08-17 | | Release date: | 2019-08-21 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | Ternary complex formation of AFN-1252 with Acinetobacter baumannii FabI and NADH: Crystallographic and biochemical studies.
Chem.Biol.Drug Des., 96, 2020
|
|
2A1W
 
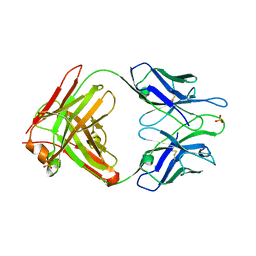 | | Anti-cocaine antibody 7.5.21, crystal form I | | Descriptor: | SULFATE ION, immunoglobulin heavy chain, immunoglobulin light chain kappa | | Authors: | Pozharski, E, Hewagama, A, Shanafelt, A, Ringe, D, Petsko, G.A. | | Deposit date: | 2005-06-21 | | Release date: | 2005-06-28 | | Last modified: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Flexibility Of Packing:
Four Crystal Forms Of An Anti-Cocaine Antibody 7.5.21
To be Published
|
|
2AI0
 
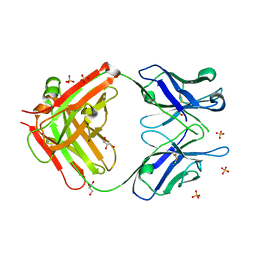 | | Anti-Cocaine Antibody 7.5.21, Crystal Form III | | Descriptor: | GLYCEROL, Immunoglobulin Heavy Chain, Immunoglobulin Light Chain kappa, ... | | Authors: | Pozharski, E, Hewagama, A, Shanafelt, A, Ringe, D, Petsko, G.A. | | Deposit date: | 2005-07-28 | | Release date: | 2005-08-09 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Flexibility of Packing: Four Crystal Forms of an Anti-Cocaine Antibody 7.5.21
To be Published
|
|
4I5I
 
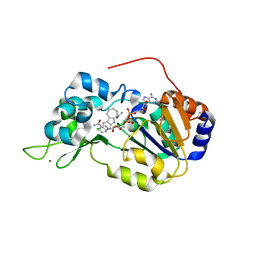 | | Crystal structure of the SIRT1 catalytic domain bound to NAD and an EX527 analog | | Descriptor: | (6S)-2-chloro-5,6,7,8,9,10-hexahydrocyclohepta[b]indole-6-carboxamide, NAD-dependent protein deacetylase sirtuin-1, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Zhao, X, Allison, D, Condon, B, Zhang, F, Gheyi, T, Zhang, A, Ashok, S, Russell, M, Macewan, I, Qian, Y, Jamison, J.A, Luz, J.G. | | Deposit date: | 2012-11-28 | | Release date: | 2013-01-23 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The 2.5 angstrom crystal structure of the SIRT1 catalytic domain bound to nicotinamide adenine dinucleotide (NAD+) and an indole (EX527 analogue) reveals a novel mechanism of histone deacetylase inhibition.
J.Med.Chem., 56, 2013
|
|
1NKW
 
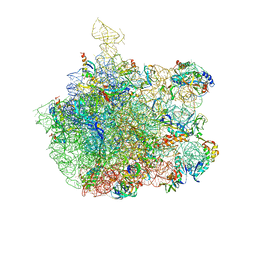 | | Crystal Structure Of The Large Ribosomal Subunit From Deinococcus Radiodurans | | Descriptor: | 23S ribosomal RNA, 50S ribosomal protein L11, 50S ribosomal protein L13, ... | | Authors: | Harms, J.M, Schluenzen, F, Zarivach, R, Bashan, A, Gat, S, Agmon, I, Bartels, H, Franceschi, F, Yonath, A. | | Deposit date: | 2003-01-05 | | Release date: | 2003-02-11 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | High resolution structure of the large ribosomal subunit from a mesophilic eubacterium
Cell(Cambridge,Mass.), 107, 2001
|
|
3OSZ
 
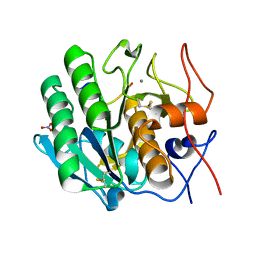 | | Crystal Structure of the complex of proteinase K with an antimicrobial nonapeptide, at 2.26 A resolution | | Descriptor: | 10-mer peptide, CALCIUM ION, NITRATE ION, ... | | Authors: | Singh, A, Sinha, M, Bhushan, A, Kaur, P, Srinivasan, A, Sharma, S, Singh, T.P. | | Deposit date: | 2010-09-10 | | Release date: | 2010-10-06 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Crystal Structure of the complex of proteinase K with an antimicrobial nonapeptide, at 2.26 A resolution
To be Published
|
|
3PIP
 
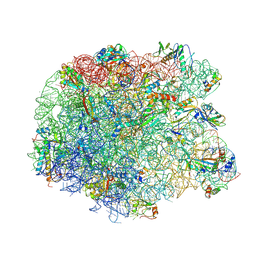 | | Crystal structure of the synergistic antibiotic pair lankamycin and lankacidin in complex with the large ribosomal subunit | | Descriptor: | 50S ribosomal protein L11, 50S ribosomal protein L13, 50S ribosomal protein L14, ... | | Authors: | Belousoff, M.J, Shapira, T, Bashan, A, Zimmerman, E, Kinashi, H, Rozenberg, H, Yonath, A. | | Deposit date: | 2010-11-07 | | Release date: | 2011-02-23 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3.45 Å) | | Cite: | Crystal structure of the synergistic antibiotic pair, lankamycin and lankacidin, in complex with the large ribosomal subunit.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
3PIO
 
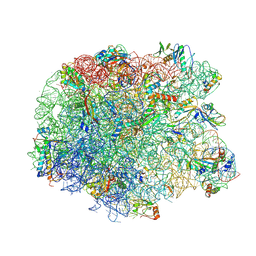 | | Crystal structure of the synergistic antibiotic pair lankamycin and lankacidin in complex with the large ribosomal subunit | | Descriptor: | 50S ribosomal protein L11, 50S ribosomal protein L13, 50S ribosomal protein L14, ... | | Authors: | Belousoff, M.J, Shapira, T, Bashan, A, Zimmerman, E, Arakawa, K, Kinashi, H, Rozenberg, H, Yonath, A. | | Deposit date: | 2010-11-07 | | Release date: | 2011-02-23 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3.2473 Å) | | Cite: | Crystal structure of the synergistic antibiotic pair, lankamycin and lankacidin, in complex with the large ribosomal subunit.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
8UDV
 
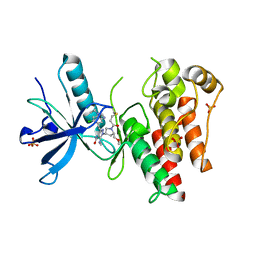 | | The X-RAY co-crystal structure of human FGFR3 V555M and Compound 17 | | Descriptor: | 1,2-ETHANEDIOL, 3-[(6-chloro-1-cyclopropyl-1H-benzimidazol-5-yl)ethynyl]-1-[(3S,5S)-5-(methoxymethyl)-1-(prop-2-enoyl)pyrrolidin-3-yl]-5-(methylamino)-1H-pyrazole-4-carboxamide, Fibroblast growth factor receptor 3, ... | | Authors: | Tyhonas, J.S, Arnold, L.D, Cox, J, Franovic, A, Gardiner, E, Grandinetti, K, Kania, R, Kanouni, T, Lardy, M, Li, C, Martin, E.S, Miller, N, Mohan, A, Murphy, E.A, Perez, M, Soroceanu, L, Timple, N, Uryu, S, Womble, S, Kaldor, S.W. | | Deposit date: | 2023-09-29 | | Release date: | 2024-02-07 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.348 Å) | | Cite: | Discovery of KIN-3248, An Irreversible, Next Generation FGFR Inhibitor for the Treatment of Advanced Tumors Harboring FGFR2 and/or FGFR3 Gene Alterations.
J.Med.Chem., 67, 2024
|
|
8UDU
 
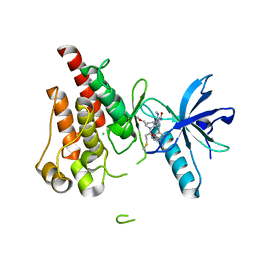 | | The X-RAY co-crystal structure of human FGFR3 and Compound 17 | | Descriptor: | 3-[(6-chloro-1-cyclopropyl-1H-benzimidazol-5-yl)ethynyl]-1-[(3S,5S)-5-(methoxymethyl)-1-(prop-2-enoyl)pyrrolidin-3-yl]-5-(methylamino)-1H-pyrazole-4-carboxamide, CHLORIDE ION, Fibroblast growth factor receptor 3 | | Authors: | Tyhonas, J.S, Arnold, L.D, Cox, J, Franovic, A, Gardiner, E, Grandinetti, K, Kania, R, Kanouni, T, Lardy, M, Li, C, Martin, E.S, Miller, N, Mohan, A, Murphy, E.A, Perez, M, Soroceanu, L, Timple, N, Uryu, S, Womble, S, Kaldor, S.W. | | Deposit date: | 2023-09-29 | | Release date: | 2024-02-07 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.737 Å) | | Cite: | Discovery of KIN-3248, An Irreversible, Next Generation FGFR Inhibitor for the Treatment of Advanced Tumors Harboring FGFR2 and/or FGFR3 Gene Alterations.
J.Med.Chem., 67, 2024
|
|
1OND
 
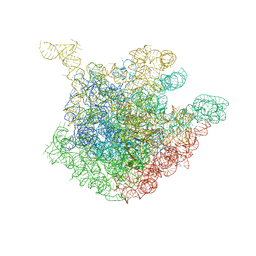 | | THE CRYSTAL STRUCTURE OF THE 50S LARGE RIBOSOMAL SUBUNIT FROM DEINOCOCCUS RADIODURANS COMPLEXED WITH TROLEANDOMYCIN MACROLIDE ANTIBIOTIC | | Descriptor: | 23S RIBOSOMAL RNA, 50S ribosomal protein L22, 50S ribosomal protein L32, ... | | Authors: | Berisio, R, Schluenzen, F, Harms, J, Bashan, A, Auerbach, T, Baram, D, Yonath, A. | | Deposit date: | 2003-02-27 | | Release date: | 2003-04-15 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Structural insight into the role of the ribosomal tunnel in cellular regulation
Nat.Struct.Biol., 10, 2003
|
|
