1T9D
 
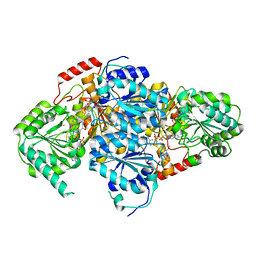 | | Crystal Structure Of Yeast Acetohydroxyacid Synthase In Complex With A Sulfonylurea Herbicide, Metsulfuron methyl | | Descriptor: | 2,5-DIMETHYL-PYRIMIDIN-4-YLAMINE, Acetolactate synthase, mitochondrial, ... | | Authors: | McCourt, J.A, Pang, S.S, Guddat, L.W, Duggleby, R.G. | | Deposit date: | 2004-05-16 | | Release date: | 2004-12-21 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Elucidating the specificity of binding of sulfonylurea herbicides to acetohydroxyacid synthase.
Biochemistry, 44, 2005
|
|
4GP1
 
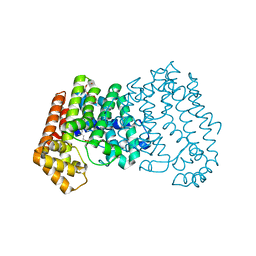 | | Crystal structure of ISOPRENOID SYNTHASE A3MSH1 (TARGET EFI-501992) from pyrobaculum calidifontis complexed with DMAPP | | Descriptor: | CHLORIDE ION, DIMETHYLALLYL DIPHOSPHATE, Polyprenyl synthetase, ... | | Authors: | Patskovsky, Y, Toro, R, Bhosle, R, Hillerich, B, Seidel, R.D, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammonds, J, Zencheck, W.D, Imker, H.J, Poulter, C.D, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2012-08-20 | | Release date: | 2012-08-29 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Crystal Structure of Isoprenoid Synthase from Pyrobaculum Calidifontis
To be Published
|
|
4H1Z
 
 | | Crystal structure of putative isomerase from Sinorhizobium meliloti, open loop conformation (target EFI-502104) | | Descriptor: | CHLORIDE ION, Enolase Q92Zs5, FORMIC ACID, ... | | Authors: | Patskovsky, Y, Toro, R, Bhosle, R, Hillerich, B, Seidel, R.D, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammonds, J, Zencheck, W.D, Imker, H.J, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2012-09-11 | | Release date: | 2012-09-26 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.012 Å) | | Cite: | Crystal Structure of Enolase Q92Zs5 (Target EFI-502104) from Sinorhizobium meliloti
To be Published
|
|
1UJL
 
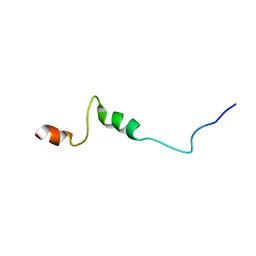 | | Solution Structure of the HERG K+ channel S5-P extracellular linker | | Descriptor: | Potassium voltage-gated channel subfamily H member 2 | | Authors: | Torres, A.M, Bansal, P.S, Sunde, M, Clarke, C.E, Bursill, J.A, Smith, D.J, Bauskin, A, Breit, S.N, Campbell, T.J, Alewood, P.F, Kuchel, P.W, Vandenberg, J.I. | | Deposit date: | 2003-08-05 | | Release date: | 2003-11-04 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Structure of the HERG K+ channel S5P extracellular linker: role of an amphipathic alpha-helix
in C-type inactivation.
J.Biol.Chem., 278, 2003
|
|
4HXA
 
 | | Crystal structure of 13D9 FAB | | Descriptor: | 13D9 FAB HEAVY CHAIN, 13D9 FAB LIGHT CHAIN | | Authors: | Johal, A.R, Jarrell, H.C, Khieu, N.H, Letts, J.A, Landry, R.C, Jachymek, W, Yang, Q, Jennings, H.J, Brisson, J.R, Evans, S.V. | | Deposit date: | 2012-11-09 | | Release date: | 2013-07-24 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | The antigen-binding site of an N-propionylated polysialic acid-specific antibody protective against group B meningococci is consistent with extended epitopes.
Glycobiology, 23, 2013
|
|
1RVK
 
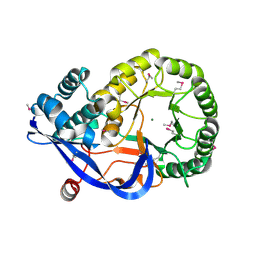 | | Crystal structure of enolase AGR_L_2751 from Agrobacterium Tumefaciens | | Descriptor: | MAGNESIUM ION, isomerase/lactonizing enzyme | | Authors: | Fedorov, A.A, Fedorov, E.V, Thirumuruhan, R, Zencheck, W, Millikin, C, Gerlt, J.A, Almo, S.C, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2003-12-14 | | Release date: | 2003-12-23 | | Last modified: | 2021-02-03 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Evolution of enzymatic activites in the Enolase superfamily: 1.7 A crystal structure of the hypothetical protein MR.GI-17937161 from Agrobacterium tumefaciens
To be Published
|
|
1RW5
 
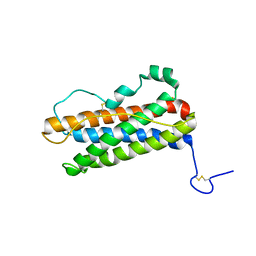 | |
1RLP
 
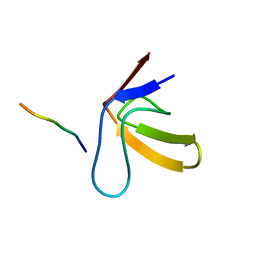 | | TWO BINDING ORIENTATIONS FOR PEPTIDES TO SRC SH3 DOMAIN: DEVELOPMENT OF A GENERAL MODEL FOR SH3-LIGAND INTERACTIONS | | Descriptor: | C-SRC TYROSINE KINASE SH3 DOMAIN, PROLINE-RICH LIGAND RLP2 (RALPPLPRY) | | Authors: | Feng, S, Chen, J.K, Yu, H, Simon, J.A, Schreiber, S.L. | | Deposit date: | 1994-10-10 | | Release date: | 1995-02-07 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Two binding orientations for peptides to the Src SH3 domain: development of a general model for SH3-ligand interactions.
Science, 266, 1994
|
|
1RRB
 
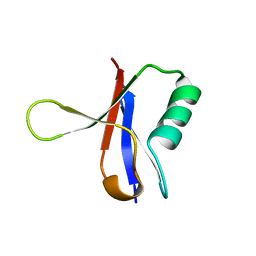 | | THE RAS-BINDING DOMAIN OF RAF-1 FROM RAT, NMR, 1 STRUCTURE | | Descriptor: | RAF PROTO-ONCOGENE SERINE/THREONINE-PROTEIN KINASE | | Authors: | Terada, T, Ito, Y, Shirouzu, M, Tateno, M, Hashimoto, K, Kigawa, T, Ebisuzaki, T, Takio, K, Shibata, T, Yokoyama, S, Smith, B.O, Laue, E.D, Cooper, J.A, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 1998-03-26 | | Release date: | 1999-03-30 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Nuclear magnetic resonance and molecular dynamics studies on the interactions of the Ras-binding domain of Raf-1 with wild-type and mutant Ras proteins.
J.Mol.Biol., 286, 1999
|
|
4JYX
 
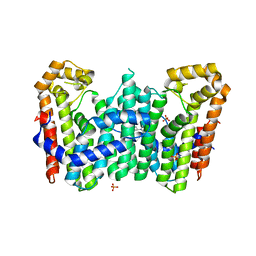 | | Crystal structure of polyprenyl synthase PATL_3739 (TARGET EFI-509195) FROM PSEUDOALTEROMONAS ATLANTICA, COMPLEX WITH INORGANIC PHOSPHATE AND AN UNKNOWN LIGAND | | Descriptor: | PHOSPHATE ION, Trans-hexaprenyltranstransferase, UNKNOWN LIGAND | | Authors: | Patskovsky, Y, Toro, R, Bhosle, R, Hillerich, B, Seidel, R.D, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammonds, J, Zencheck, W.D, Imker, H.J, Al Obaidi, N, Stead, M, Love, J, Poulter, C.D, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2013-04-01 | | Release date: | 2013-04-10 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structure of Isoprenoid Synthase Patl_3739 from Pseudoalteromonas Atlantica
To be Published
|
|
1RMK
 
 | | Solution structure of conotoxin MrVIB | | Descriptor: | Mu-O-conotoxin MrVIB | | Authors: | Daly, N.L, Ekberg, J.A, Thomas, L, Adams, D.J, Lewis, R.J, Craik, D.J. | | Deposit date: | 2003-11-28 | | Release date: | 2004-09-07 | | Last modified: | 2022-03-02 | | Method: | SOLUTION NMR | | Cite: | Structures of muO-conotoxins from Conus marmoreus. Inhibitors of tetrodotoxin (TTX)-sensitive and TTX-resistant sodium channels in mammalian sensory neurons
J.Biol.Chem., 279, 2004
|
|
1RN9
 
 | | A SHORT LEXITROPSIN THAT RECOGNIZES THE DNA MINOR GROOVE AT 5'-ACTAGT-3': UNDERSTANDING THE ROLE OF ISOPROPYL-THIAZOLE | | Descriptor: | 5'-D(*CP*GP*AP*CP*TP*AP*GP*TP*CP*G)-3' | | Authors: | Anthony, N.G, Johnston, B.F, Khalaf, A.I, Mackay, S.P, Parkinson, J.A, Suckling, C.J, Waigh, R.D. | | Deposit date: | 2003-12-01 | | Release date: | 2004-08-24 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Short Lexitropsin that Recognizes the DNA Minor Groove at 5'-ACTAGT-3': Understanding the Role of Isopropyl-thiazole.
J.Am.Chem.Soc., 126, 2004
|
|
4K2K
 
 | | Crystal structure of Staphylococcal nuclease variant Delta+PHS L25A/L36A/I92A at cryogenic temperature | | Descriptor: | CALCIUM ION, THYMIDINE-3',5'-DIPHOSPHATE, Thermonuclease | | Authors: | Caro, J.A, Schlessman, J.L, Heroux, A, Garcia-Moreno E, B. | | Deposit date: | 2013-04-09 | | Release date: | 2013-04-24 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Pressure effects on proteins
To be Published
|
|
4GYP
 
 | | Crystal structure of the heterotetrameric complex of GlucD and GlucDRP from E. coli K-12 MG1655 (EFI TARGET EFI-506058) | | Descriptor: | CITRIC ACID, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Lukk, T, Ghasempur, S, Imker, H.J, Gerlt, J.A, Nair, S.K, Enzyme Function Initiative (EFI) | | Deposit date: | 2012-09-05 | | Release date: | 2012-09-12 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Glucarate dehydratase and its related protein from Escherichia coli form a heterotetrameric complex.
to be published
|
|
4H2H
 
 | | Crystal structure of an enolase (mandalate racemase subgroup, target EFI-502101) from Pelagibaca bermudensis htcc2601, with bound mg and l-4-hydroxyproline betaine (betonicine) | | Descriptor: | (2S,4R)-4-hydroxy-1,1-dimethylpyrrolidinium-2-carboxylate, (4S)-2-METHYL-2,4-PENTANEDIOL, IODIDE ION, ... | | Authors: | Vetting, M.W, Morisco, L.L, Wasserman, S.R, Sojitra, S, Imker, H.J, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2012-09-12 | | Release date: | 2012-10-10 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Discovery of new enzymes and metabolic pathways by using structure and genome context.
Nature, 502, 2013
|
|
4HG7
 
 | | Crystal Structure of an MDM2/Nutlin-3a complex | | Descriptor: | 4-({(4S,5R)-4,5-bis(4-chlorophenyl)-2-[4-methoxy-2-(propan-2-yloxy)phenyl]-4,5-dihydro-1H-imidazol-1-yl}carbonyl)piperazin-2-one, E3 ubiquitin-protein ligase Mdm2, SULFATE ION | | Authors: | Noble, M.E.M, Anil, B, Riedinger, C, Endicott, J.A. | | Deposit date: | 2012-10-07 | | Release date: | 2013-07-31 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The structure of an MDM2-Nutlin-3a complex solved by the use of a validated MDM2 surface-entropy reduction mutant.
Acta Crystallogr.,Sect.D, 69, 2013
|
|
1UUG
 
 | | ESCHERICHIA COLI URACIL-DNA GLYCOSYLASE:INHIBITOR COMPLEX WITH WILD-TYPE UDG AND WILD-TYPE UGI | | Descriptor: | URACIL-DNA GLYCOSYLASE, URACIL-DNA GLYCOSYLASE INHIBITOR | | Authors: | Mol, C.D, Arvai, A.S, Putnam, C.D, Tainer, J.A. | | Deposit date: | 1998-10-31 | | Release date: | 1999-03-25 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Protein mimicry of DNA from crystal structures of the uracil-DNA glycosylase inhibitor protein and its complex with Escherichia coli uracil-DNA glycosylase
J.Mol.Biol., 287, 1999
|
|
4GVX
 
 | | Crystal structure of a short chain dehydrogenase homolog (target EFI-505321) from burkholderia multivorans, with bound NADP and L-fucose | | Descriptor: | 1,2-ETHANEDIOL, 3-oxoacyl-[acyl-carrier protein] reductase, CHLORIDE ION, ... | | Authors: | Vetting, M.W, Hobbs, M.E, Morisco, L.L, Wasserman, S.R, Sojitra, S, Imker, H.J, Raushel, F.M, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2012-08-31 | | Release date: | 2012-10-03 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.499 Å) | | Cite: | Crystal structure of a short chain dehydrogenase homolog (target EFI-505321) from burkholderia multivorans, with bound NADP and L-fucose
To be Published
|
|
1UZZ
 
 | | Erythrina cristagalli bound to N-linked oligosaccharide and lactose | | Descriptor: | CALCIUM ION, GLYCEROL, Lectin, ... | | Authors: | Turton, K, Natesh, R, Thiyagarajan, N, Chaddock, J.A, Acharya, K.R. | | Deposit date: | 2004-03-20 | | Release date: | 2004-06-22 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.13 Å) | | Cite: | Crystal structures of Erythrina cristagalli lectin with bound N-linked oligosaccharide and lactose.
Glycobiology, 14, 2004
|
|
4JZ5
 
 | |
4K1X
 
 | | Ferredoxin-NADP(H) Reductase mutant with Ala 266 replaced by Tyr (A266Y) and residues 267-272 deleted. | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, NADPH:ferredoxin reductase, SULFATE ION | | Authors: | Bortolotti, A, Sanchez-Azqueta, A, Maya, C.M, Velazquez-Campoy, A, Hermoso, J.A, Cortez, N. | | Deposit date: | 2013-04-06 | | Release date: | 2013-11-13 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The C-terminal extension of bacterial flavodoxin-reductases: Involvement in the hydride transfer mechanism from the coenzyme.
Biochim.Biophys.Acta, 1837, 2013
|
|
4K1W
 
 | | Crystal structure of the A314P mutant of mannonate dehydratase from novosphingobium aromaticivorans complexed with mg and d-mannonate | | Descriptor: | 1,3-PROPANDIOL, CARBON DIOXIDE, CARBONATE ION, ... | | Authors: | Fedorov, A.A, Fedorov, E.V, Wichelecki, D, Gerlt, J.A, Almo, S.C. | | Deposit date: | 2013-04-06 | | Release date: | 2014-04-09 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.646 Å) | | Cite: | Crystal structure of the A314P mutant of mannonate
dehydratase from novosphingobium aromaticivorans complexed with mg
and d-mannonate
To be Published
|
|
4K74
 
 | |
4K8L
 
 | | Crystal structure of a putative 4-hydroxyproline epimerase/3-hydroxyproline dehydratse from the soil bacterium ochrobacterium anthropi, target efi-506495, disordered loops | | Descriptor: | Proline racemase | | Authors: | Vetting, M.W, Toro, R, Bhosle, R, Al Obaidi, N.F, Morisco, L.L, Wasserman, S.R, Sojitra, S, Stead, M, Washington, E, Scott Glen, A, Chowdhury, S, Evens, B, Hammonds, J, Hillerich, B, Love, J, Seidel, R.D, Imker, H.J, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2013-04-18 | | Release date: | 2013-05-01 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of a putative 4-hydroxyproline epimerase/3-hydroxyproline dehydratse from the soil bacterium ochrobacterium anthropi, target efi-506495, disordered loops
To be Published
|
|
1T6Q
 
 | | Nickel Superoxide Dismutase (NiSOD) CN-treated Apo Structure | | Descriptor: | Superoxide dismutase [Ni] | | Authors: | Barondeau, D.P, Kassmann, C.J, Bruns, C.K, Tainer, J.A, Getzoff, E.D. | | Deposit date: | 2004-05-07 | | Release date: | 2004-07-13 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Nickel superoxide dismutase structure and mechanism.
Biochemistry, 43, 2004
|
|
