1INL
 
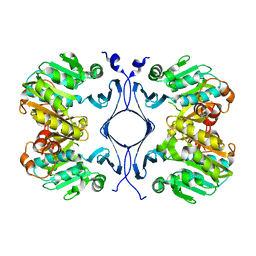 | | Crystal Structure of Spermidine Synthase from Thermotoga Maritima | | Descriptor: | Spermidine synthase | | Authors: | Korolev, S, Skarina, T, Ikeguchi, Y, Pegg, A.E, Joachimiak, A, Edwards, A, Savchenko, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2001-05-14 | | Release date: | 2001-11-21 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The crystal structure of spermidine synthase with a multisubstrate adduct inhibitor.
Nat.Struct.Biol., 9, 2002
|
|
1L7A
 
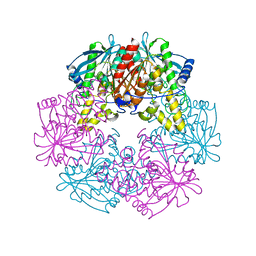 | |
5VD6
 
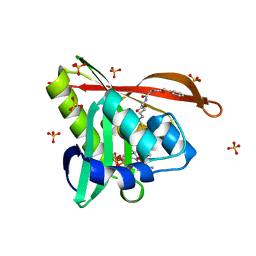 | | Crystal structure of a GNAT superfamily acetyltransferase PA4794 in complex with bisubstrate analog 6 | | Descriptor: | (3R,5S,9R,23S)-1-[(2R,3S,4R,5R)-5-(6-amino-9H-purin-9-yl)-4-hydroxy-3-(phosphonooxy)tetrahydrofuran-2-yl]-3,5,9-trihydroxy-8,8-dimethyl-10,14-dioxo-23-({[(phenylacetyl)amino]acetyl}amino)-2,4,6-trioxa-18-thia-11,15-diaza-3,5-diphosphatetracosan-24-oic acid 3,5-dioxide (non-preferred name), SULFATE ION, acetyltransferase PA4794 | | Authors: | Majorek, K.A, Joachimiak, A, Minor, W, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2017-04-01 | | Release date: | 2017-07-26 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Generating enzyme and radical-mediated bisubstrates as tools for investigating Gcn5-related N-acetyltransferases.
FEBS Lett., 591, 2017
|
|
1KR4
 
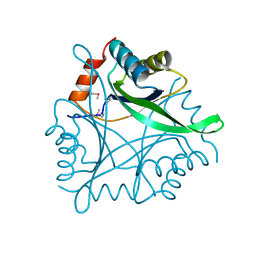 | | Structure Genomics, Protein TM1056, cutA | | Descriptor: | Protein TM1056, cutA | | Authors: | Savchenko, A, Zhang, R, Joachimiak, A, Edwards, A, Akarina, T, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2002-01-08 | | Release date: | 2002-08-14 | | Last modified: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | X-ray crystal structure of CutA from Thermotoga maritima at 1.4 A resolution.
Proteins, 54, 2004
|
|
1JQ3
 
 | | Crystal Structure of Spermidine Synthase in Complex with Transition State Analogue AdoDATO | | Descriptor: | S-ADENOSYL-1,8-DIAMINO-3-THIOOCTANE, Spermidine synthase | | Authors: | Korolev, S, Ikeguchi, Y, Skarina, T, Beasley, S, Edwards, A, Joachimiak, A, Pegg, A.E, Savchenko, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2001-08-03 | | Release date: | 2001-11-21 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The crystal structure of spermidine synthase with a multisubstrate adduct inhibitor.
Nat.Struct.Biol., 9, 2002
|
|
1K7K
 
 | | crystal structure of RdgB- inosine triphosphate pyrophosphatase from E. coli | | Descriptor: | Hypothetical protein yggV | | Authors: | Sanishvili, R, Joachimiak, A, Edwards, A, Savchenko, A, Skarina, T, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2001-10-19 | | Release date: | 2002-08-14 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Molecular basis of the antimutagenic activity of the house-cleaning inosine triphosphate pyrophosphatase RdgB from Escherichia coli.
J.Mol.Biol., 374, 2007
|
|
1KYH
 
 | | Structure of Bacillus subtilis YxkO, a Member of the UPF0031 Family and a Putative Kinase | | Descriptor: | Hypothetical 29.9 kDa protein in SIGY-CYDD intergenic region | | Authors: | Zhang, R, Dementieva, I, Vinokour, E, Collart, F, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2002-02-04 | | Release date: | 2002-08-14 | | Last modified: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure of Bacillus subtilis YXKO--a member of the UPF0031 family and a putative kinase.
J.Struct.Biol., 139, 2002
|
|
1L6R
 
 | | Crystal Structure of Thermoplasma acidophilum 0175 (APC0014) | | Descriptor: | CALCIUM ION, FORMIC ACID, HYPOTHETICAL PROTEIN TA0175 | | Authors: | Kim, Y, Joachimiak, A, Edwards, A.M, Xu, X, Pennycooke, M, Gu, J, Cheung, F, Christendat, D, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2002-03-13 | | Release date: | 2003-01-21 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structure- and function-based characterization of a new phosphoglycolate phosphatase from Thermoplasma acidophilum.
J.Biol.Chem., 279, 2004
|
|
1M6Y
 
 | | Crystal Structure Analysis of TM0872, a Putative SAM-dependent Methyltransferase, Complexed with SAH | | Descriptor: | S-ADENOSYL-L-HOMOCYSTEINE, S-adenosyl-methyltransferase mraW, SULFATE ION | | Authors: | Miller, D.J, Anderson, W.F, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2002-07-17 | | Release date: | 2003-01-28 | | Last modified: | 2016-03-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal complexes of a predicted S-adenosylmethionine-dependent methyltransferase reveal a typical AdoMet binding domain and a substrate recognition domain
Protein Sci., 12, 2003
|
|
1N2X
 
 | | Crystal Structure Analysis of TM0872, a Putative SAM-dependent Methyltransferase, Complexed with SAM | | Descriptor: | S-ADENOSYLMETHIONINE, S-adenosyl-methyltransferase mraW, SULFATE ION | | Authors: | Miller, D.J, Anderson, W.F, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2002-10-24 | | Release date: | 2003-01-28 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal complexes of a predicted S-adenosylmethionine-dependent methyltransferase reveal a typical AdoMet binding domain and a substrate recognition domain
Protein Sci., 12, 2003
|
|
3VDH
 
 | | Crystal structure of PbGH5A, a glycoside hydrolase family 5 enzyme from Prevotella bryantii B14 | | Descriptor: | B-1,4-endoglucanase, CHLORIDE ION | | Authors: | Stogios, P.J, Evdokimova, E, Egorova, O, Yim, V, Joachimiak, A, Edwards, A.M, Savchenko, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2012-01-05 | | Release date: | 2012-01-18 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Structure-Function Analysis of a Mixed-linkage beta-Glucanase/Xyloglucanase from the Key Ruminal Bacteroidetes Prevotella bryantii B14.
J.Biol.Chem., 291, 2016
|
|
3VCX
 
 | | Crystal structure of a putative glyoxalase/bleomycin resistance protein from Rhodopseudomonas palustris CGA009 | | Descriptor: | Glyoxalase/Bleomycin resistance protein/dioxygenase domain, TETRAETHYLENE GLYCOL | | Authors: | Stogios, P.J, Chang, C, Evdokimova, E, Egorova, O, Joachimiak, A, Edwards, A.M, Savchenko, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2012-01-04 | | Release date: | 2012-01-18 | | Last modified: | 2012-01-25 | | Method: | X-RAY DIFFRACTION (1.39 Å) | | Cite: | Crystal structure of a putative glyoxalase/bleomycin resistance protein from Rhodopseudomonas palustris CGA009
To be Published
|
|
1U60
 
 | | MCSG APC5046 Probable glutaminase ybaS | | Descriptor: | 1,2-ETHANEDIOL, FORMIC ACID, Probable glutaminase ybaS | | Authors: | Chang, C, Cuff, M.E, Joachimiak, A, Savchenko, A, Edwards, A, Skarina, T, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2004-07-28 | | Release date: | 2004-09-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | Functional and structural characterization of four glutaminases from Escherichia coli and Bacillus subtilis.
Biochemistry, 47, 2008
|
|
2ZC2
 
 | | Crystal structure of DnaD-like replication protein from Streptococcus mutans UA159, gi 24377835, residues 127-199 | | Descriptor: | DnaD-like replication protein, ZINC ION | | Authors: | Duke, N.E.C, Clancy, S, Duggan, E, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2007-11-02 | | Release date: | 2007-12-25 | | Last modified: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structure of DnaD-like replication protein from Streptococcus mutans UA159.
To be Published
|
|
3B7H
 
 | |
3B4S
 
 | |
3B49
 
 | |
3B85
 
 | |
3B4Q
 
 | |
3B79
 
 | |
3BJD
 
 | | Crystal structure of putative 3-oxoacyl-(acyl-carrier-protein) synthase from Pseudomonas aeruginosa | | Descriptor: | 1,2-ETHANEDIOL, NICKEL (II) ION, Putative 3-oxoacyl-(acyl-carrier-protein) synthase | | Authors: | Osipiuk, J, Evdokimova, E, Kudritska, M, Savchenko, A, Edwards, A.M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2007-12-03 | | Release date: | 2007-12-11 | | Last modified: | 2017-10-25 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal structure of putative 3-oxoacyl-(acyl-carrier-protein) synthase from Pseudomonas aeruginosa.
To be Published
|
|
3B33
 
 | |
3BJN
 
 | | Crystal structure of C-terminal domain of putative transcriptional regulator from Vibrio cholerae, targeted domain 79-240 | | Descriptor: | CHLORIDE ION, Transcriptional regulator, putative | | Authors: | Chang, C, Volkart, L, Clancy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2007-12-04 | | Release date: | 2007-12-11 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystal structure of C-terminal domain of putative transcriptional regulator from Vibrio cholerae.
To be Published
|
|
3BEE
 
 | | Crystal structure of putative YfrE protein from Vibrio parahaemolyticus | | Descriptor: | 1,2-ETHANEDIOL, Putative YfrE protein | | Authors: | Chang, C, Hatzos, C, Abdullah, J, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2007-11-16 | | Release date: | 2007-12-04 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Crystal structure of putative YfrE protein from Vibrio parahaemolyticus.
To be Published
|
|
3BHG
 
 | | Crystal structure of adenylosuccinate lyase from Legionella pneumophila | | Descriptor: | Adenylosuccinate lyase, GLYCEROL, SULFATE ION | | Authors: | Chang, C, Li, H, Freeman, L, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2007-11-28 | | Release date: | 2007-12-04 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of adenylosuccinate lyase from Legionella pneumophila.
To be Published
|
|
