1KZV
 
 | |
2CYM
 
 | | EFFECTS OF AMINO ACID SUBSTITUTION ON THREE-DIMENSIONAL STRUCTURE: AN X-RAY ANALYSIS OF CYTOCHROME C3 FROM DESULFOVIBRIO VULGARIS HILDENBOROUGH AT 2 ANGSTROMS RESOLUTION | | Descriptor: | CYTOCHROME C3, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Morimoto, Y, Tani, T, Okumura, H, Higuchi, Y, Yasuoka, N. | | Deposit date: | 1993-09-29 | | Release date: | 1994-04-30 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Effects of amino acid substitution on three-dimensional structure: an X-ray analysis of cytochrome c3 from Desulfovibrio vulgaris Hildenborough at 2 A resolution.
J.Biochem.(Tokyo), 110, 1991
|
|
1M9G
 
 | | Solution structure of G16A-MNEI, a structural mutant of single chain monellin MNEI | | Descriptor: | Monellin chain B and Monellin chain A | | Authors: | Spadaccini, R, Trabucco, F, Saviano, G, Picone, D, Crescenzi, O, Tancredi, T, Temussi, P.A. | | Deposit date: | 2002-07-29 | | Release date: | 2003-06-10 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | The Mechanism of Interaction of Sweet Proteins with the T1R2-T1R3 Receptor: Evidence from the Solution Structure of G16A-MNEI
J.MOL.BIOL., 328, 2003
|
|
1TRM
 
 | |
3AU4
 
 | |
1VFP
 
 | |
3VNH
 
 | | Crystal Structure of Keap1 Soaked with Synthetic Small Molecular | | Descriptor: | 2-(3-((3-(5-(furan-2-yl)-1,3,4-oxadiazol-2-yl)ureido)methyl)phenoxy)acetic acid, Kelch-like ECH-associated protein 1 | | Authors: | Kunishima, N, Tanaka, T, Satoh, M, Saburi, H. | | Deposit date: | 2012-01-12 | | Release date: | 2013-01-16 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structure of Keap1 Soaked with Synthetic Small Molecular
To be Published
|
|
3VNG
 
 | | Crystal Structure of Keap1 in Complex with Synthetic Small Molecular based on a co-crystallization | | Descriptor: | 2-(3-((3-(5-(furan-2-yl)-1,3,4-oxadiazol-2-yl)ureido)methyl)phenoxy)acetic acid, Kelch-like ECH-associated protein 1 | | Authors: | Kunishima, N, Tanaka, T, Satoh, M, Saburi, H. | | Deposit date: | 2012-01-12 | | Release date: | 2013-01-16 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structure of Keap1 in Complex with Synthetic Small Molecular based on a co-crystallization
To be Published
|
|
4L28
 
 | | Crystal structure of delta516-525 human cystathionine beta-synthase D444N mutant containing C-terminal 6xHis tag | | Descriptor: | Cystathionine beta-synthase, PROTOPORPHYRIN IX CONTAINING FE, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Ereno, J, Majtan, T, Oyenarte, I, Kraus, J.P, Martinez, L.A. | | Deposit date: | 2013-06-04 | | Release date: | 2013-09-18 | | Last modified: | 2013-10-16 | | Method: | X-RAY DIFFRACTION (2.626 Å) | | Cite: | Structural basis of regulation and oligomerization of human cystathionine beta-synthase, the central enzyme of transsulfuration.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
3WN8
 
 | | Crystal Structure of Collagen-Model Peptide, (POG)3-PRG-(POG)4 | | Descriptor: | collagen-like peptide | | Authors: | Okuyama, K, Haga, M, Noguchi, K, Tanaka, T. | | Deposit date: | 2013-12-06 | | Release date: | 2014-08-13 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Preferred side-chain conformation of arginine residues in a triple-helical structure.
Biopolymers, 101, 2014
|
|
3W59
 
 | |
3W58
 
 | |
4L0D
 
 | | Crystal structure of delta516-525 human cystathionine beta-synthase containing C-terminal 6xHis-tag | | Descriptor: | Cystathionine beta-synthase, PROTOPORPHYRIN IX CONTAINING FE, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Ereno, J, Majtan, T, Oyenarte, I, Kraus, J.P, Martinez-Cruz, L.A. | | Deposit date: | 2013-05-31 | | Release date: | 2013-09-18 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.97 Å) | | Cite: | Structural basis of regulation and oligomerization of human cystathionine beta-synthase, the central enzyme of transsulfuration.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
4KFZ
 
 | | Crystal structure of LMO2 and anti-LMO2 VH complex | | Descriptor: | Anti-LMO2 VH, LMO-2, ZINC ION | | Authors: | Sewell, H, Tanaka, T, El Omari, K, Cruz-Migoni, A, Mancini, E.J, Fuentes-Fernandez, N, Chambers, J, Rabbitts, T.H. | | Deposit date: | 2013-04-28 | | Release date: | 2014-01-22 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Conformational flexibility of the oncogenic protein LMO2 primes the formation of the multi-protein transcription complex.
Sci Rep, 4, 2014
|
|
4L27
 
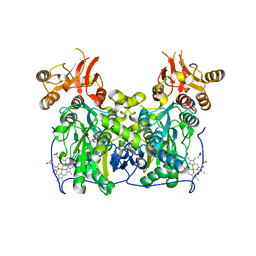 | | Crystal structure of delta1-39 and delta516-525 human cystathionine beta-synthase D444N mutant containing C-terminal 6xHis tag | | Descriptor: | Cystathionine beta-synthase, PROTOPORPHYRIN IX CONTAINING FE, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Ereno, J, Majtan, T, Oyenarte, I, Kraus, J.P, Martinez, L.A. | | Deposit date: | 2013-06-04 | | Release date: | 2013-09-18 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.391 Å) | | Cite: | Structural basis of regulation and oligomerization of human cystathionine beta-synthase, the central enzyme of transsulfuration.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
4L3V
 
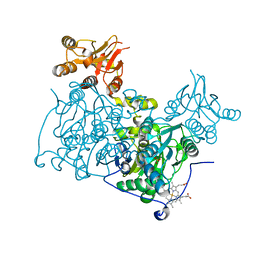 | | Crystal structure of delta516-525 human cystathionine beta-synthase | | Descriptor: | Cystathionine beta-synthase, PROTOPORPHYRIN IX CONTAINING FE, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Ereno, J, Majtan, T, Oyenarte, I, Kraus, J.P, Martinez, L.A. | | Deposit date: | 2013-06-07 | | Release date: | 2013-09-18 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.628 Å) | | Cite: | Structural basis of regulation and oligomerization of human cystathionine beta-synthase, the central enzyme of transsulfuration.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
4APW
 
 | | Alp12 filament structure | | Descriptor: | ALP12 | | Authors: | Popp, D, Narita, A, Lee, L.J, Ghoshdastider, U, Xue, B, Srinivasan, R, Balasubramanian, M.K, Tanaka, T, Robinson, R.C. | | Deposit date: | 2012-04-06 | | Release date: | 2012-05-16 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (19.700001 Å) | | Cite: | Novel Actin-Like Filament Structure from Clostridium Tetani.
J.Biol.Chem., 287, 2012
|
|
5YAE
 
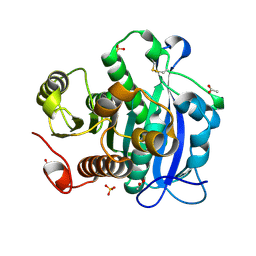 | | Ferulic acid esterase from Streptomyces cinnamoneus at 2.4 A resolution | | Descriptor: | ACETATE ION, Esterase, SULFATE ION | | Authors: | Tamura, H, Uraji, M, Mizohata, E, Ogawa, K, Inoue, T, Hatanaka, T. | | Deposit date: | 2017-08-31 | | Release date: | 2017-12-06 | | Last modified: | 2018-01-31 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Loop of Streptomyces Feruloyl Esterase Plays an Important Role in the Enzyme's Catalyzing the Release of Ferulic Acid from Biomass.
Appl. Environ. Microbiol., 84, 2018
|
|
5YAL
 
 | | Ferulic acid esterase from Streptomyces cinnamoneus at 1.5 A resolution | | Descriptor: | ACETATE ION, Esterase, GLYCEROL, ... | | Authors: | Tamura, H, Uraji, M, Mizohata, E, Ogawa, K, Inoue, T, Hatanaka, T. | | Deposit date: | 2017-09-01 | | Release date: | 2017-12-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Loop of Streptomyces Feruloyl Esterase Plays an Important Role in the Enzyme's Catalyzing the Release of Ferulic Acid from Biomass.
Appl. Environ. Microbiol., 84, 2018
|
|
3WSU
 
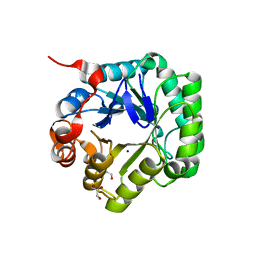 | | Crystal structure of beta-mannanase from Streptomyces thermolilacinus | | Descriptor: | Beta-mannanase, GLYCEROL, SODIUM ION | | Authors: | Kumagai, Y, Yamashita, K, Okuyama, M, Hatanaka, T, Yao, M, Kimura, A. | | Deposit date: | 2014-03-26 | | Release date: | 2015-05-20 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The loop structure of Actinomycete glycoside hydrolase family 5 mannanases governs substrate recognition
Febs J., 282, 2015
|
|
3WHO
 
 | | X-ray-Crystallographic Structure of an RNase Po1 Exhibiting Anti-tumor Activity | | Descriptor: | Guanyl-specific ribonuclease Po1 | | Authors: | Kobayashi, H, Katsurtani, T, Hara, Y, Motoyoshi, N, Itagaki, T, Akita, F, Higashiura, A, Yamada, Y, Suzuki, M, Inokuchi, N. | | Deposit date: | 2013-08-30 | | Release date: | 2014-07-02 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | X-ray crystallographic structure of RNase Po1 that exhibits anti-tumor activity.
Biol.Pharm.Bull., 37, 2014
|
|
6IBI
 
 | | Copper binding protein from Laetisaria arvalis (LaX325) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Auxiliary activity CAZyme, COPPER (II) ION, ... | | Authors: | Frandsen, K.E.H, Tandrup, T, Labourel, A, Haon, M, Berrin, J.-G, Lo Leggio, L. | | Deposit date: | 2018-11-30 | | Release date: | 2019-11-13 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | A fungal family of lytic polysaccharide monooxygenase-like copper proteins.
Nat.Chem.Biol., 16, 2020
|
|
6IBJ
 
 | | Copper binding protein from Laetisaria arvalis (LaX325) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Auxiliary activity CAZyme, COPPER (II) ION | | Authors: | Frandsen, K.E.H, Tandrup, T, Labourel, A, Haon, M, Berrin, J.-G, Lo Leggio, L. | | Deposit date: | 2018-11-30 | | Release date: | 2019-11-13 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | A fungal family of lytic polysaccharide monooxygenase-like copper proteins.
Nat.Chem.Biol., 16, 2020
|
|
6IBH
 
 | | Copper binding protein from Laetisaria arvalis (LaX325) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Auxiliary activity CAZyme, COPPER (II) ION, ... | | Authors: | Frandsen, K.E.H, Tandrup, T, Labourel, A, Haon, M, Berrin, J.-G, Lo Leggio, L. | | Deposit date: | 2018-11-30 | | Release date: | 2019-11-13 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | A fungal family of lytic polysaccharide monooxygenase-like copper proteins.
Nat.Chem.Biol., 16, 2020
|
|
3WHB
 
 | |
